With the emergence of SARS-CoV-2 variants in late 2020, we saw an opportunity to modify this test. We also wanted to make it more specific to neutralizing antibodies (the ones that provide some protection from disease) and thought that using our platform we could assess the humoral immune response (both from natural infection and from vaccination) against several variants simultaneously.
We wanted to build a test that could be used to help determine someone's risks for re-infection or breakthrough infections based on variants that have emerged.

Image Credit: Mediantone/Shutterstock.com
Despite new viral variants being discovered, we still currently have no rapid ways of assessing variants. Why is this and why is it important to be able to quickly determine their presence?
The gold standard method for assessing the effectiveness of neutralizing antibodies against SARS-CoV-2 variants is through live virus microneutralization of plaque reduction neutralization tests. These tests are extremely important in understanding the ability of SARS-CoV-2 variants to evade immunity; however, they are time-consuming to perform, require biosafety level 3 facilities/highly trained personnel, and are thus non-accessible in many places around the world.
We think our test can supplement conventional tests and address some of their limitations – it is accessible, easy-to-operate, and rapid.
Can you describe how you designed your latest test? How does it work?
Our CoVariant-SCAN (COVID-19 Variant Spike-ACE2–Competitive Antibody Neutralization) assay evaluates the ability of host neutralizing antibodies to block the pathologic interaction between variants of viral spike proteins and the human ACE2 receptor within 1 hour from a drop of plasma. This assay is constructed by inkjet-printing a portion of the spike protein (RBD) from each variant onto a “non-fouling” poly(oligo ethylene glycol methyl ether methacrylate) (POEGMA) coating. This non-fouling coating eliminates nearly all non-specific protein adsorption and cellular adhesion, leading to a very sensitive test.
Nearby, fluorescently labeled human ACE2 is inkjet-printed upon a dissolvable trehalose pad. When a sample without neutralizing antibodies is added to CoVariant-SCAN, fluorescently labeled ACE2 dissolves from the POEMGA brush and binds to RBD capture sites, leading to a high fluorescence signal. In the presence of potential neutralizing antibodies, the RBD-ACE2 interaction can be partially or completely blocked, resulting in a decrease in fluorescence signal.
We demonstrate the multiplexing capability of CoVariant-SCAN by simultaneously assessing the neutralizing activity against wild type, B.1.1.7 (Alpha), B.1.351 (Beta), P.1 (Gamma), and B.1.617.2 (Delta) variants.
How sensitive is your test at determining which COVID-19 variant is present?
Our test does not seek to necessarily determine which COVID-19 variant somebody is infected with. Rather, the goal is to determine how protected you might be from infection from a given variant after vaccination or natural infection. It can also be used to identify individuals who develop a weak humoral immune response and thus may benefit from monoclonal antibody therapies if they become infected or exposed.
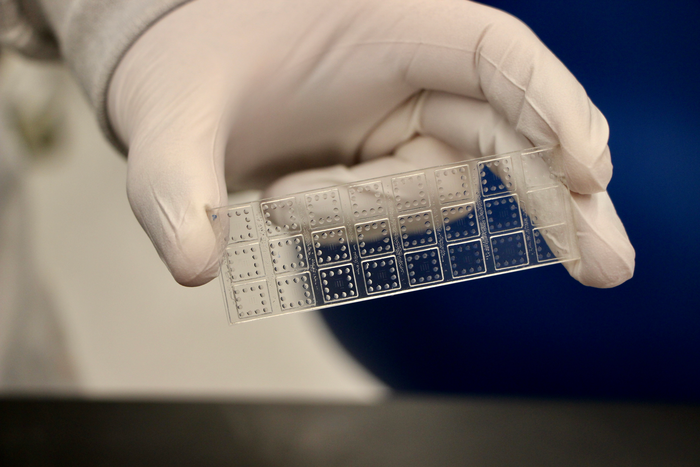
Image Credit: Duke University
What are the benefits of your test compared to others that have been developed?
Compared to the conventional tests (as discussed previously), our test is more rapid and much easier to operate. Other tests have been developed that use a similar operating principle, for example, https://www.genscript.com/covid-19-detection-cpass.html.
However, this assay only measures neutralizing antibodies against the initial SARS-CoV-2 strain, whereas ours can assess neutralizing antibodies against several SARS-COV-2 variants simultaneously from a single sample.
How could your research be used to determine how well a patient is protected from emerging COVID-19 variants?
We envision our test could be used to do just that. An individual could test their blood for neutralizing antibodies and gain some information about how well they may be protected from certain variants that are included in the test.
Luckily, our test is relatively easy to modify. When the delta variant emerged, we demonstrated that we could rapidly incorporate it into our platform. We are now in the process of doing the same thing for Omicron.
Could your test also determine which monoclonal antibody treatment would work best for any given patient?
Absolutely, it could be helpful for that type of application. In the paper, we showed that the Regerneon cocktail of antibodies retained activity against all variants tested. Other groups have shown that certain antibodies are not as effective against certain variants.
Our test could be used to screen for monoclonal antibodies to rapidly assess the effectiveness against a given variant and could also be used to potentially identify patients who may benefit from receiving prophylactic monoclonal antibody therapy (or treatment early in disease), for example, if they have a compromised immune system.
Image Credit: Cristian Storto/Shutterstock.com
Do you believe that with continued research into COVID-19 variants, we can be better equipped to deal with new ones if they emerge?
Yes. In my opinion, robust genomic surveillance systems and widespread vaccination are our best tools for being equipped to deal with SARS-CoV-2 variants.
We need to be able to identify new variants quickly, determine the functional impact of the mutations, and continue to protect ourselves via vaccination.
How are you beginning to streamline your technique and what advantages will this have?
We hope to get to a point where we can manufacture these tests at scale using more sustainable manufacturing methods. We are also working on simplifying the testing process even further using microfluidics (similar to the format used here: https://www.science.org/doi/10.1126/sciadv.abg4901).
Once these are accomplished, it will allow us to hopefully further validate the devices and gain approval for the tests to be used in the broader population prospectively.
What are your next steps in your research?
First, working on the scalable manufacturing process and microfluidic implementation (as described above). Second, with the emergence of the Omicron variant, we are rapidly trying to adapt the platform to include its RBD protein. With that, we hope to be able to assess whether or not Omicron is able to evade the humoral immune response.
Where can readers find more information?
The link to this publication is here: https://www.science.org/doi/10.1126/sciadv.abl7682
The link to our previous publication, detailing our microfluidic chip and portable detector is here: https://www.science.org/doi/10.1126/sciadv.abg4901
Other relevant publications from our lab, including our Ebola test (https://stm.sciencemag.org/content/13/588/eabd9696) and breast cancer test (https://www.nature.com/articles/s41523-021-00290-0).
About Jacob Heggestad
Jacob Heggestad received his Bachelor's degree in chemical engineering from Northwestern University in 2017.
He is currently a Ph.D. Candidate in Biomedical Engineering in the Chilkoti lab at Duke University, where he is conducting research on point-of-care diagnostics.
About Professor Ashutosh Chilkoti
Ashutosh Chilkoti is the Alan L. Kaganov Professor and the Chair of the Department of Biomedical Engineering at Duke University. His areas of research include genetically encoded materials, biointerface science, and nonfouling polymer brush surfaces for point-of-care diagnostics. He is a member of the National Academy of Inventors. He is also the founder of five start-up companies.