
 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
Immunological responses induced by our body in response to any pathogenic attack lead to the development of immunological memory, which further protects against future reinfection. In the lungs, MBCs protect against any reinfection in the respiratory system; however, the biology of these cells is not well understood.
About the study
The present study aims to determine whether respiratory organ MBCs occupy specific tissue niches within the lung mucosa and if these cells bear special transcriptional programs that permit their survival in the lung airways.
Further, the researchers were interested in identifying the presence of distinct MBCs subsets that may arise upon infection. The same outcomes were also investigated for lymphoid organs.
The researchers intranasally infected the Aicda-CreERT2 Rosa26-EYFP mouse strain (Aid-EYFP mice) with five plaque-forming units (PSUs) of influenza. Comparatively, K18-hACE2 mice expressing the human angiotensin-converting enzyme 2 (ACE2) receptor were infected with 2.103 PFU of SARS-CoV-2, for determining MBCs across these models.
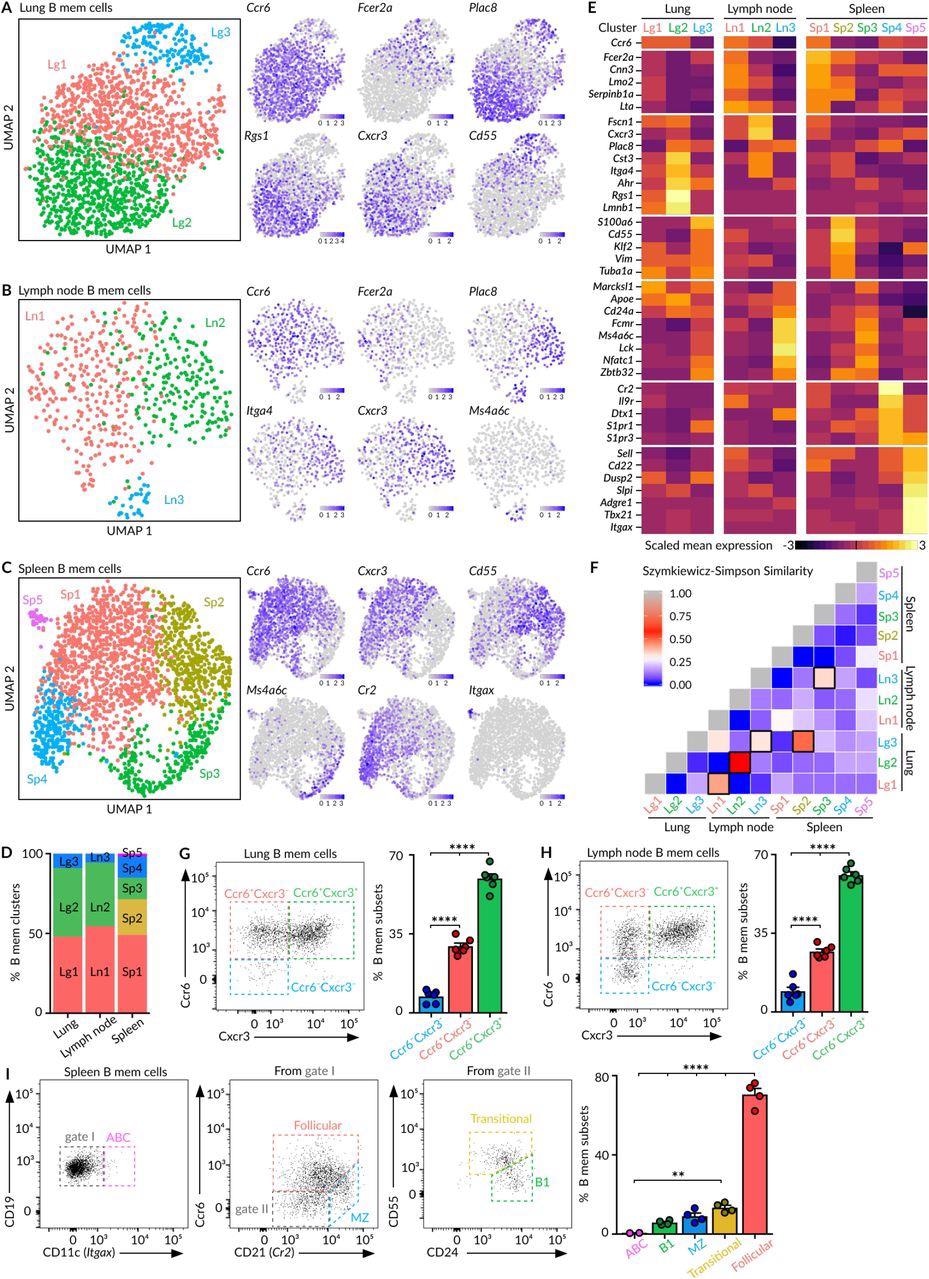 Heterogeneity of MBCs (A-C) UMAP projections of MBCs in lungs (A), mediastinal lymph node (B) and spleen (C), colored by subpopulation. Feature plots display the expression of indicated marker genes in MBCs laid out in the UMAP representation. Scale: normalized UMI counts. (D) Bar chart showing the relative proportions of MBC subpopulations within each tissue. (E) Heatmaps exhibiting marker gene expression for each tissue subpopulation. Colour: Scaled mean expression. (F) Szymkiewicz-Simpson similarity matrix for pairwise comparisons among subpopulations from different tissues. Black rectangles indicate values higher than 0.32. (G-H) Flow cytometry plots showing the presence of Ccr6-Cxcr3-, Ccr6+Cxcr3- and Ccr6+Cxcr3+ MBC subsets in the CD19+ YFP+CD38+GL7- population from lungs (G) and lymph nodes (H). (I) Flow cytometry plots showing the gating strategy for spleen MBC subsets according to the expression of Cd11c, Ccr6, Cd21, Cd24 and Cd55. In panels G to I, bar charts show the quantification of one representative experiment out of three, mean ± s.e.m. Each dot represents one mouse. One-way Anova test: **p<0.01 and ****p<0.0001.
Heterogeneity of MBCs (A-C) UMAP projections of MBCs in lungs (A), mediastinal lymph node (B) and spleen (C), colored by subpopulation. Feature plots display the expression of indicated marker genes in MBCs laid out in the UMAP representation. Scale: normalized UMI counts. (D) Bar chart showing the relative proportions of MBC subpopulations within each tissue. (E) Heatmaps exhibiting marker gene expression for each tissue subpopulation. Colour: Scaled mean expression. (F) Szymkiewicz-Simpson similarity matrix for pairwise comparisons among subpopulations from different tissues. Black rectangles indicate values higher than 0.32. (G-H) Flow cytometry plots showing the presence of Ccr6-Cxcr3-, Ccr6+Cxcr3- and Ccr6+Cxcr3+ MBC subsets in the CD19+ YFP+CD38+GL7- population from lungs (G) and lymph nodes (H). (I) Flow cytometry plots showing the gating strategy for spleen MBC subsets according to the expression of Cd11c, Ccr6, Cd21, Cd24 and Cd55. In panels G to I, bar charts show the quantification of one representative experiment out of three, mean ± s.e.m. Each dot represents one mouse. One-way Anova test: **p<0.01 and ****p<0.0001.
Study findings
The study results revealed previously unidentified and high levels of heterogeneity in secondary lymphoid organ MBCs. In particular, the spleen constitutes a distinct segment for a minimum of five MBC subsets including follicular, transitional, B1-like, marginal zone, and age-associated (ABC) MBCs that cohabitate this organ upon resolution of respiratory disease infection.
The follicular MBC cluster (Sp1) displayed certain levels of class-switching towards (immunoglobulin G) IgG isotypes, whereas immunoglobulin M (IgM)+ was expressed in transitional (Sp2), B1-like (Sp3), marginal zone (Sp4), and ABC (Sp5) MBCs.
In the lungs, the MBCs formed clusters adjacent to lung bronchi, which allows for the rapid encounter of pathogens during re-infection and thus ensures the first layer of protection at the tissue level directly.
The single-cell RNA sequencing (scRNA-seq) analysis showed that the MBC pool in the lungs and draining lymph nodes are identical to each other at the level of cluster composition and subsequent B cell receptor (BCR) sequences, thereby indicating association in the origin of these two organs. The results of this analysis also revealed the three distinct clusters of MBCs residing in the lungs and lymph nodes.
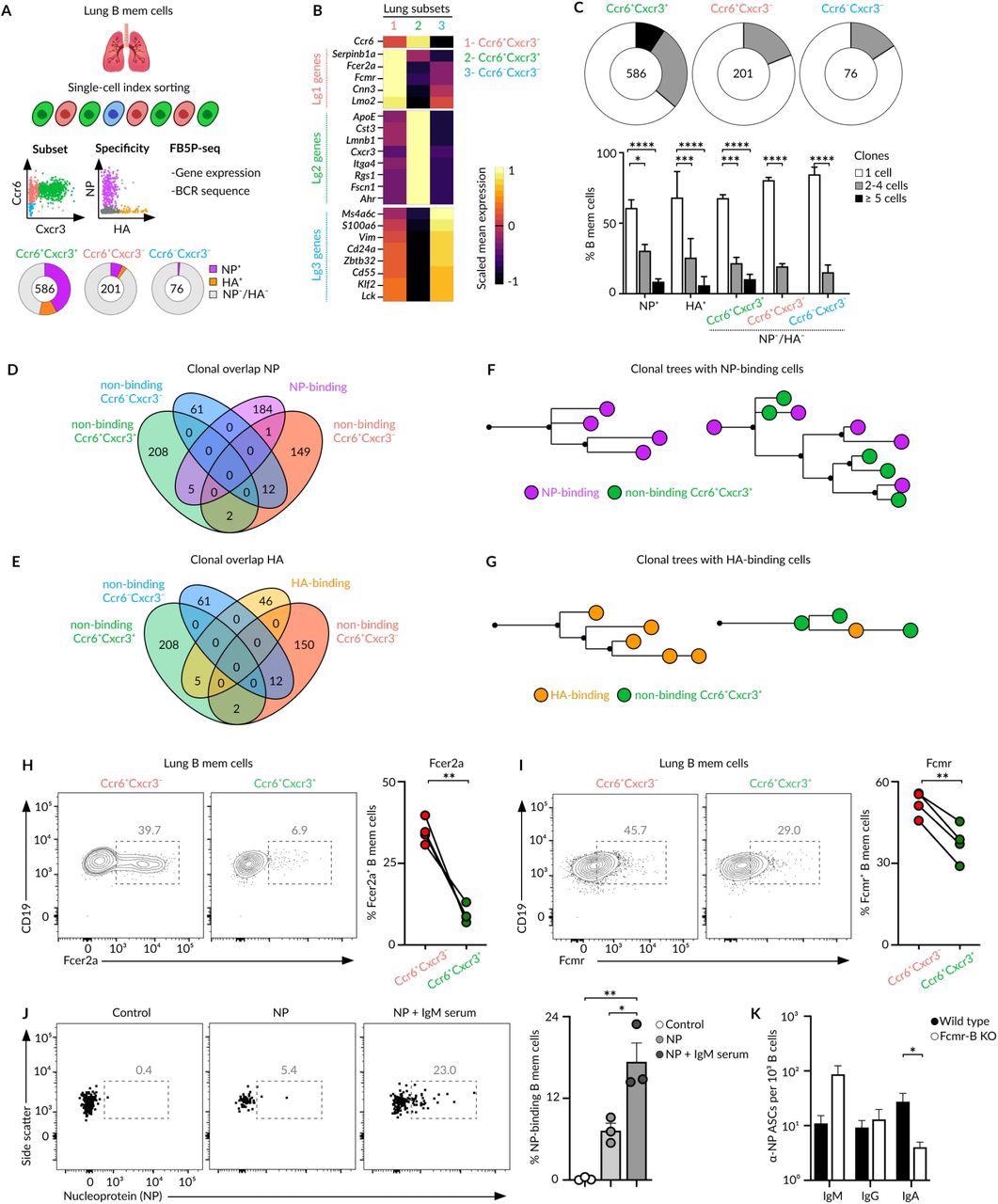 Bona fide and bystander MBCs (A) Overview of the index cell sorting and FB5P-seq experimental workflow. Information of Ccr6/Cxcr3 expression and HA/NP binding was recorded for each lung MBC sorted. Quantification of HA/NP binding cells in different memory subsets is shown in pie charts. (B) Heatmap showing the expression of marker genes from Lg1, Lg2 and Lg3 clusters (10X dataset) by Ccr6+Cxcr3-, Ccr6+Cxcr3+ and Ccr6-Cxcr3- subsets. Colour: Scaled mean expression. (C) Pie charts displaying the percentage of cells comprising single-cell, 2-4 cell, or ≥4 cell clones for each subset. Bar charts showing clonal size in NP/HA binding and non-binding memory cells, two-way Anova test. (D-E) Venn diagrams showing the clonal overlap among non-binding cells from Ccr6+Cxcr3-, Ccr6+Cxcr3+ and Ccr6-Cxcr3- subsets and NP-binding (D) or HA-binding (E) cells. (F-G) Trees showing phylogenetic relationships of IgH and IgK sequences from clones containing NP-binding (F) and HA-binding (G) cells. (H-I) Flow cytometry plots showing the expression of Fcer2a (H) and Fcmr (I) by Ccr6+Cxcr3- and Ccr6+Cxcr3+ MBC subsets, paired t-test. (J) Flow cytometry plots showing NP binding to Ccr6+Cxcr3- lung MBCs after incubation with media, NP or NP-IgM from immune sera, t-test. (K) Enumeration of IgM, IgG and IgA ASCs measured by ELISPOT in lungs of chimeric mice with a wild type or Fcmr-deficient B cell compartment. Mice were infected with Influenza PR8, challenged with influenza X31 after 40 days and sacrificed 4 days later Anova test. In all panels, each dot represents one mouse, mean ± s.e.m, *p<0.01.*p<0.05, ***p<0.001 and ****p<0.0001.
Bona fide and bystander MBCs (A) Overview of the index cell sorting and FB5P-seq experimental workflow. Information of Ccr6/Cxcr3 expression and HA/NP binding was recorded for each lung MBC sorted. Quantification of HA/NP binding cells in different memory subsets is shown in pie charts. (B) Heatmap showing the expression of marker genes from Lg1, Lg2 and Lg3 clusters (10X dataset) by Ccr6+Cxcr3-, Ccr6+Cxcr3+ and Ccr6-Cxcr3- subsets. Colour: Scaled mean expression. (C) Pie charts displaying the percentage of cells comprising single-cell, 2-4 cell, or ≥4 cell clones for each subset. Bar charts showing clonal size in NP/HA binding and non-binding memory cells, two-way Anova test. (D-E) Venn diagrams showing the clonal overlap among non-binding cells from Ccr6+Cxcr3-, Ccr6+Cxcr3+ and Ccr6-Cxcr3- subsets and NP-binding (D) or HA-binding (E) cells. (F-G) Trees showing phylogenetic relationships of IgH and IgK sequences from clones containing NP-binding (F) and HA-binding (G) cells. (H-I) Flow cytometry plots showing the expression of Fcer2a (H) and Fcmr (I) by Ccr6+Cxcr3- and Ccr6+Cxcr3+ MBC subsets, paired t-test. (J) Flow cytometry plots showing NP binding to Ccr6+Cxcr3- lung MBCs after incubation with media, NP or NP-IgM from immune sera, t-test. (K) Enumeration of IgM, IgG and IgA ASCs measured by ELISPOT in lungs of chimeric mice with a wild type or Fcmr-deficient B cell compartment. Mice were infected with Influenza PR8, challenged with influenza X31 after 40 days and sacrificed 4 days later Anova test. In all panels, each dot represents one mouse, mean ± s.e.m, *p<0.01.*p<0.05, ***p<0.001 and ****p<0.0001.
The lungs and lymph nodes represent the MBC subsets with the unique expression pattern of CXCR3 and CCR6 receptors. Among these subsets, the smallest subset is comprised of innate-like MBCs, which exclusively express IgM and lack CXCR3 and CCR6 expression.
These cells were localized at the peripheral part of the lungs per-bronchial B-cell clusters and do not manifest specificity for viral antigens. However, their gene expression and B-cell receptor (BCR) sequences exhibited resemblance to splenic B1-like MBCs.
The CXCR3- and CXCR3+ MBCs subsets share the peri-bronchial niche in the lung mucosa and arise from germinal centers based on their CD40-L dependence and somatic hypermutation levels. The CXCR3- and CXCR3+ MBCs subsets undergo extensive class-switching towards IgG subclasses.
During ex vivo infections, these subsets differentiate into plasma cells, rather than into germinal center cells, and produce similar levels of IgG. Overall, these results indicated that CXCR3- and CXCR3+ have distinct memory populations and do not segregate as per fate but generate through divergent mechanisms.
The bona fide MBCs constitute pathogen-specific CXCR3+, which is recruited actively during re-infection. The CXCR3- subset represented bystander MBCs with no specificity for pathogen-derived antigens.
SARS-CoV-2 infection also induces bonafide and bystander MBC subsets with contrasting antigen specificities. The benefit of memory subset formation is still not known, despite the mechanisms and as bystander cells represent half (50%) of the MBC pool generated during infection. The researchers suggest that the low and high-affinity MBCs within the bona fide subset can help patients fight against future virus attacks, mutating variants, and different viruses from the same family.
Conclusions
These results challenged the notion that germinal centers only work as “machines” that produce high-affinity bona fide MBCs. Instead, permissive selection mechanisms operate these reactions, expand the diversity of the initial B-cell repertoire, and give rise to bystander MBCs.
Thus, diverse transcriptional programs in MBCs are not linked to specific effector fates but rather to divergent strategies of the immune system to simultaneously provide rapid protection from reinfection while diversifying the initial B-cell repertoire.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Gregoire, C., Spinelli, L., Villazala-Merino, S., et al. (2021). Viral infection engenders bona fide and bystander lung memory B cell subsets through permissive selection. bioRxiv. doi:10.1101/2021.12.14.472614. https://www.biorxiv.org/content/10.1101/2021.12.14.472614v1.
- Peer reviewed and published scientific report.
Gregoire, Claude, Lionel Spinelli, Sergio Villazala-Merino, Laurine Gil, María Pía Holgado, Myriam Moussa, Chuang Dong, et al. 2022. “Viral Infection Engenders Bona Fide and Bystander Subsets of Lung-Resident Memory B Cells through a Permissive Mechanism.” Immunity 55 (7): 1216-1233.e9. https://doi.org/10.1016/j.immuni.2022.06.002. https://www.cell.com/immunity/fulltext/S1074-7613(22)00240-0.