
 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
Major histocompatibility complex class I (MHC-I) molecules play an important role in mediating adaptive immune responses to viral infections. These molecules are heterodimers of a glycosylated transmembrane heavy chain and the small protein β2-microglobulin (β2m).
MHC-I is synthesized and assembled in the endoplasmic reticulum (ER), after which they are loaded with peptides from the turnover of cellular proteins. Peptide loading is achieved by the peptide loading complex (PLC), ER chaperone proteins ERp57 and calreticulin, as well as the peptide editor tapasin. Thereafter, the peptide-MHC-I complex moves to the surface of cells, where non-self-peptide-expressing cells are identified and killed by cytotoxic T-lymphocytes (CTLs).
Viruses, including the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that is responsible for the ongoing coronavirus disease 2019 (COVID-19) pandemic, can downregulate the expression of MHC-I, either by transcriptional downregulation of MHC-I genes or post-translational inhibition of MHC-I. Many viruses can also encode pore-forming viral proteins or viroporins that can disrupt ion homeostasis by integrating into the host membranes.
SARS-CoV-2 is a single-stranded, positive-sense enveloped ribonucleic acid (RNA) virus that encodes several open reading frames (ORFs) which, in turn, encodes structural, non-structural, and accessory proteins. Previous studies have reported that SARS-CoV-2 accessory proteins can reduce the surface expression of a specific MHC-I allele, human leukocyte antigen (HLA)-A*02:01.
About the study
The current study involved infection of Vero E6 cells by SARS-CoV-2, followed by the determination of viral titers using the plaque assay. Thereafter, immunoblotting, flow cytometry, co-immunoprecipitation, and immunofluorescence assays were carried out. A quantitative reverse transcription-polymerase chain reaction (qRT-PCR) assay was conducted using the RNA extracted from the cells, followed by complement DNA (cDNA) synthesis.
Metabolic labeling and immunoprecipitation were achieved through the use of cells that expressed inducible ORF7a. Structural analysis of ORF7a, human β2m, and HLA-A2/β2m/peptide complex, as well as molecular dynamics simulations and binding free energy analysis, were also conducted.
Study findings
The expression of MHC-I was reduced by 30-40% in SARS-CoV-2-infected cells as compared to uninfected cells. The three proteins associated with the downregulation of MHC-I expression included ORF3a, ORF7a, and ORF8. Both ORF7a and ORF8 were hypothesized to specifically affect MHC-I expression, while ORF3a was hypothesized to impair protein trafficking through the secretory pathway.
The expression of ORF3a led to fragmentation of the Golgi apparatus, which subsequently led to downregulation of surface MHC-I since the trafficking of glycoproteins to the cell surface was blocked.
ORF7a was observed to slow the rate of export of MHC-I from the ER by four hours as a result of its interference with the antigen processing pathway through its interaction with the MHC-I heavy chain (HC). Interaction of HC-ORF7a was more stable with higher total energy as compared to HC- β2m. However, an excess of β2m could overcome the action of ORF7a.
Analysis of an ORF7a with mutated ER-retrieval sequence, which was referred to as ORF7a-ARA, reported that ORF7a localized primarily in the Golgi. Induction of ORF7a also reduced MHC-I expression by 40%, while no such observation was made for ORF7a-ARA induction. Along with the reduction in MHC-I expression, ORF7a also led to a decrease in antigen presentation by reduction of surface HLA-A2.
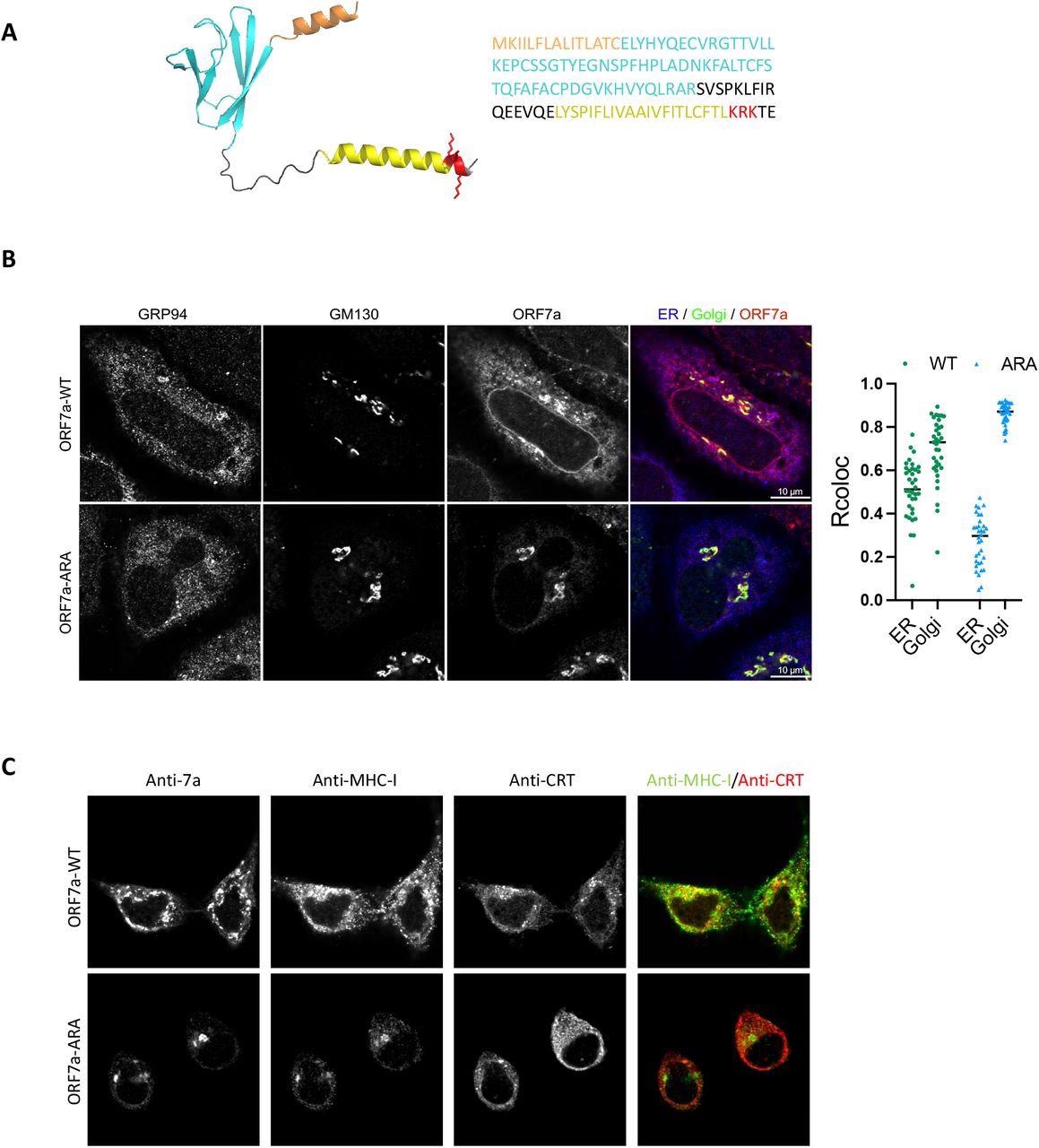
Localization of ORF7a to the ER determines retention of MHC-I. (A) A structural model of the full-length ORF7a as generated by AlphaFold (left) and amino acid sequence of the protein (right). Color coding indicates functional features of the protein-orange, ER-targeting signal sequence; cyan, immunoglobulin-like fold; yellow, transmembrane domain; red, the ER-retrieval motif (KRK) in the cytosolic C-terminal tail with the side chains shown. (B) Localization of ORF7a by confocal analysis. HeLaM cells were transfected with plasmids encoding ORF7a-WT or ORF7a-ARA and 24 h post-transfection cells were fixed and stained with GRP94 (ER, blue), GM130 (Golgi, green), and ORF7a, red (scale bars, 10 μm) to determine colocalization (n= 35). (C) Localization of MHC-I in the presence of ORF7a-WT or ORF7a-ARA was determined by transfecting HeLaM cells with plasmids encoding ORF7a-WT or ORF7a-ARA for 24 h followed by a confocal analysis of ORF7a, MHC-I (green) and calreticulin (ER, red). Scale bars, 10 μm.
Conclusions
The current study determined that both ORF7a and ORF3a could manipulate MHC-I levels post-translationally by distinct mechanisms. Both of these proteins cause a delay in MHC-I trafficking, as well as impaired antigen presentation.
Further research is required to understand the mechanisms that contribute to COVID-19 pathogenicity for the development of novel therapeutics, as well as for the understanding of any related emergent pathogens in the future.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Arshad, N., Laurent-Rolle, M., Ahmed, W. S., et al. (2022). SARS-CoV-2 accessory proteins ORF7a and ORF3a use distinct mechanisms to downregulate MHC-I surface expression. bioRxiv. doi:10.1101/2022.05.17.492198. https://www.biorxiv.org/content/10.1101/2022.05.17.492198v1.
- Peer reviewed and published scientific report.
Arshad, Najla, Maudry Laurent-Rolle, Wesam S. Ahmed, Jack Chun-Chieh Hsu, Susan M. Mitchell, Joanna Pawlak, Debrup Sengupta, Kabir H. Biswas, and Peter Cresswell. 2022. “SARS-CoV-2 Accessory Proteins ORF7a and ORF3a Use Distinct Mechanisms to Down-Regulate MHC-I Surface Expression.” Proceedings of the National Academy of Sciences 120 (1). https://doi.org/10.1073/pnas.2208525120. https://www.pnas.org/doi/10.1073/pnas.2208525120.