The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is still causing hundreds of thousands of infections around the world each week. Cases are mostly asymptomatic, but also there are a significant number of severe or even lethal infections. Already, in just over six months, the disease has claimed over 609,000 lives worldwide, from over 14.7 million cases.
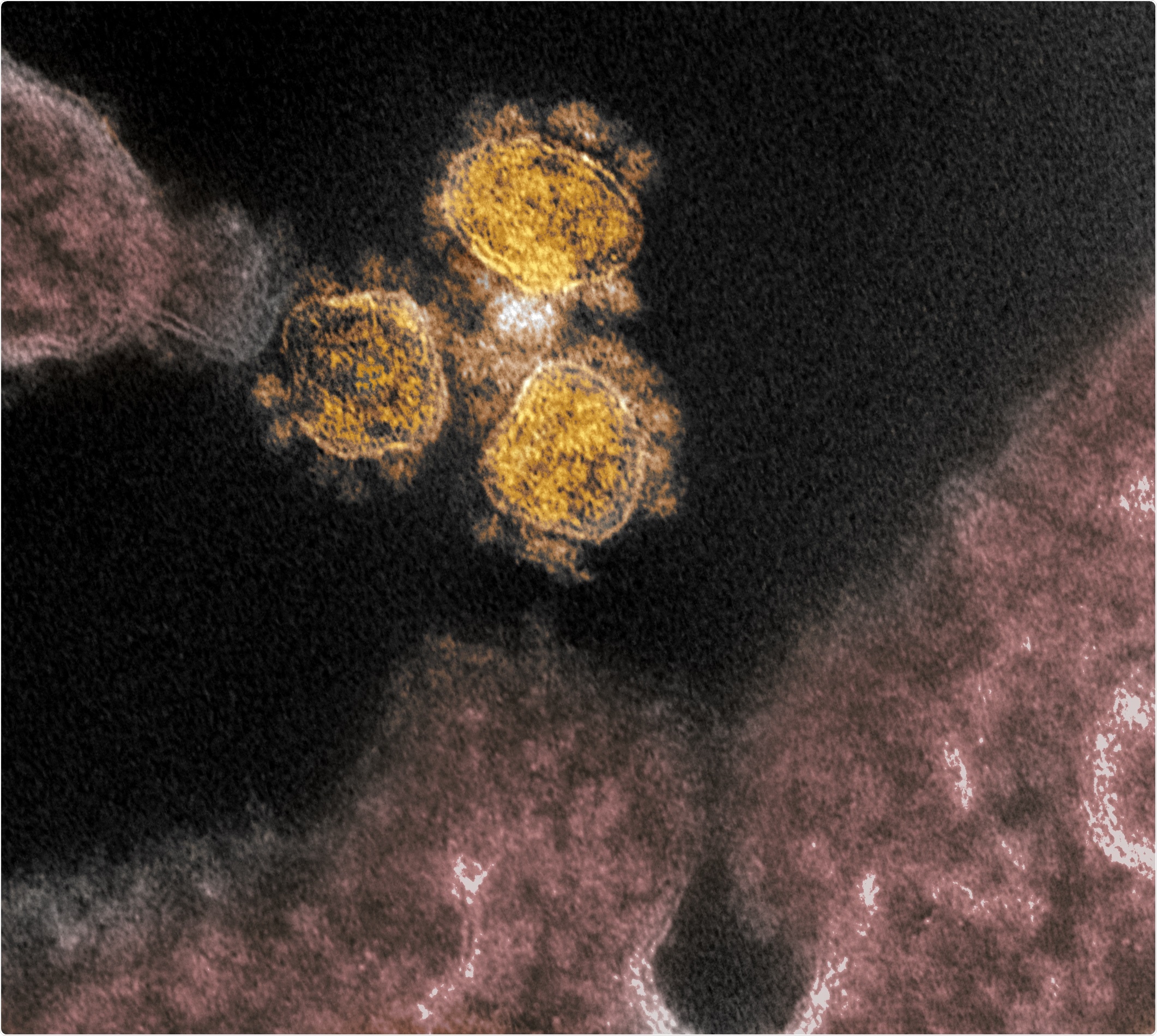
Novel Coronavirus SARS-CoV-2 - This transmission electron microscope image shows SARS-CoV-2, the virus that causes COVID-19, isolated from a patient in the U.S. Virus particles (round gold objects) are shown emerging from the surface of cells cultured in the lab. The spikes on the outer edge of the virus particles give coronaviruses their name, crown-like. Image captured and colorized at NIAID's Rocky Mountain Laboratories (RML) in Hamilton, Montana. Credit: NIAID
Among the most frustrating features of COVID-19 disease has been the inability to forecast which patients will progress to severe disease, require intensive care unit (ICU), and die from the disease. Evidence is accumulating that among the most important factors is an imbalance between the innate and adaptive arms of the immune response. Severely ill COVID-19 patients develop a steep fall in the number of lymphocytes, while chemicals that reflect inflammation register an increase. These chemicals include C-Reactive Protein (CRP), IL-1β, TNF-α, IL-8, and IL-6.
A second clue is the efficacy of corticosteroids and the cytokine inhibitor tocilizumab, which inhibits the IL-6 receptor. Earlier research suggested that an exuberant inflammatory response was responsible for progressive disease and multi-organ damage in COVID-19. However, currently, it is thought that there is also an abnormality in the way the innate and adaptive arms of the immune response balance each other.
Discrepancies are evident in the reports of the patient immune profile from different studies, which may be due to sampling at different times or in patients with a different clinical presentation. Now, a new study published on the preprint server medRxiv* aims to analyze the immune response over time, using multi-omics in the study of a single cell.
The Study: Multi-Omics Over Time
The researchers from Yale University examined 18 paired samples collected at 2 different time points from all but 2 patients with progressive illness who died before a second sample was available. The samples contained peripheral blood mononuclear cells (PBMCs), obtained from 10 COVID-19 patients with a range of outcomes, in addition to 13 samples from healthy controls who were age-matched.
Among the 10 patients, 4 had progressive disease and expired, with 6 having stable disease culminating in discharge. Tocilizumab was used for 8/10 patients. All patients were similar when the baseline and timeline characteristics were compared.
They employed different approaches, including 5' single-cell RNA sequencing for gene expression (GEX), Cellular Indexing of Transcriptomes and Epitopes by sequencing (CITE-seq), and B and T cell receptor repertoire analysis. They matched the multi-omics results with clinical and laboratory data, including the viral load and plasma level of various cytokines, taken over a period of time.
Type 1 IFN Response
They found that all cell types show a continually changing type-1 interferon (IFN) response, which lessens over time. It tends to increase from the first to the second sample in progressive patients. Both IFN-activated CD8 T cells and ISGs are increased as part of this response.
IL-10 and MHC-II Levels
IL-10 expression also follows the same trend, declining in stable patients but increased in progressive patients. MHC-II levels are reduced, and this combination ensures a reduction in inflammation-related organ damage but also reduces the body's ability to clear the viral response. They also found that this correlates with a reduction in viral load and that both are more obvious in COVID-19 patients with progressive disease.
AREG Expression
Secondly, they found that monocytes are expressing anti-inflammatory cytokines in progressive COVID-19. This includes expression of amphiregulin (AREG), which binds to epidermal growth factor receptor, and is involved in wound repair as well as a resolution of inflammation. It is higher in viral lung infections, and mice with SARS-CoV infection develop severe lung disease in the presence of AREG. It is also induced by ISG expression in response to type 1 IFN signaling and has been shown to be increased in the PBMCs of COVID-19 patients as well.
ISG Expression
And thirdly, they found that IFN-activated CD8 cells expressing IFN-stimulated genes (ISG), are also a characteristic of extreme illness. Effector T cells and naïve T cells are increased and decreased, respectively, in progressive patients compared to stable patients. The former also have higher plasmablast number and dividing T and natural killer cells as well compared to controls.
Activated T Cells and Preterminally Exhausted T Cells
Another finding was of higher activated T cells with dual expression of HLADR*, and CD38* markers represent T cells stimulated by an acute viral infection. These are found in COVID-19 patients, both stable and progressive, compared to controls. However, T cells expressing MKI67 were found at higher levels, especially in the late phase of COVID-19. Activated T cells in progressive patients also express ISGs as well as a unique set of co-inhibitory receptors (LAG3 and TIM3) markers that promote T cell exhaustion. They also display higher expression of ISGs and increased secretion of pro-inflammatory or cytotoxic cytokines.
Uniquely, the researchers found a T cell phenotype between the already described progenitor or stem-like exhausted state and the terminally exhausted state, which is termed a pre-terminally exhausted state. Dividing T cells expressed more terminally exhausted markers in progressive patients, as well as type 1 IFN response genes. In short, the current study suggests that type 1 IFN promotes preterminal T cell exhaustion.
The reasons for this premature T cell exhaustion are unclear since COVID-19 patients are not exposed to viral antigens for a very long period. LAG3 binds to MHC-II, and the interruption of this binding could be one of the important factors for the disordered immunopathology.
Out of Sync
Overall, there was a disconnect between the two arms of the immune system, which the researchers say resembles immunoparalysis. The monocyte profile showed a shift from classical monocyte to monocyte-derived suppressor cell (MDSC)-like profile, favoring disease resolution. This could be the result of a strong type 1-IFN response.
Difference in Response to Tocilizumab
Tocilizumab shows variation in effects across the range of cell types, perhaps because of the difference in the way IL6R and IL6ST are expressed. Overall, the composite IL-6 score shows a decline in all patients on tocilizumab, but for one. This also markedly reduces the expression of specific genes across most cell types. Higher expression of these genes, S100A8, and S100A9, are characteristic of severe COVID-19.
Possible Explanations for Study Observations
It is thought that these genes enhance the expression of IL-6, which in turn increases the expression of these genes, resulting in increased inflammation. This could help account for the beneficial effect of tocilizumab, via inhibition of these genes.
The IL6R :IL6ST ratio is high in monocytes and other myeloid cells, unlike all other types of cells. Prior research suggests that this could be an important factor in deciding how the body responds to IL-6, either via classical anti-inflammatory pathways involving myeloid cells, or pro-inflammatory trans pathways involving other cells. This could explain why different cell types react differently to tocilizumab.
Moreover, the distribution of CD8 T cells is altered, while the primary B cells are prominent in the immune response, reflected by the high number of unmutated IGHG B cell numbers. Mutated clones may be the result of the stimulation of pre-existing memory B cells.
Finally, platelet genes are expressed at higher levels in monocytes, indicating the presence of platelet-monocyte aggregates, in progressive patients. This could be due to the elevated IL-6 and IL-8 levels. The activated platelets may elicit anti-inflammatory macrophage shifts and also increase the expression of IL-10.
Implications and Conclusion
The researchers explain that this potentially immunosuppressive shift, while the viral load is still high, could spell the difference between progressive and stable disease by disrupting the antiviral immune response. The desynchronized adaptive/innate immune response, with higher expression of ISGs and inflammatory molecules like AREG, and cytokine IL-10.
The study concludes: "Overall, our comprehensive immune profiling underscores the desynchronized innate and adaptive immune interaction in progressive COVID-19, which may lead to delayed virus clearance. This high-resolution understanding of the immune cell profiles underlying severe COVID-19 will enhance our ability to develop immunomodulatory therapeutic approaches to prevent progression in COVID-19 patients."

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources