Researchers from Sweden and Brazil have identified a novel mutation in a strain of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) obtained from a patient in Stockholm, Sweden, in late April.
The mutation is located at the surface of the S1 subunit of the SARS-CoV-2 spike protein, the main surface structure the virus uses to gain entry to host cells. The S1 subunit contains the binding domain for the host cell membrane receptor angiotensin-converting enzyme 2 (ACE2).
The study also found that of four near-complete genomes assembled from infected patients, three belonged to one of the genetic groups that the Public Health Agency of Sweden had previously reported as having declined in prevalence by late April.
The team warns that further investigation is needed to ensure that the spread of different SARS-CoV-2 strains is properly characterized.
A pre-print version of the paper is available on the server bioRxiv*, while the article undergoes peer review.
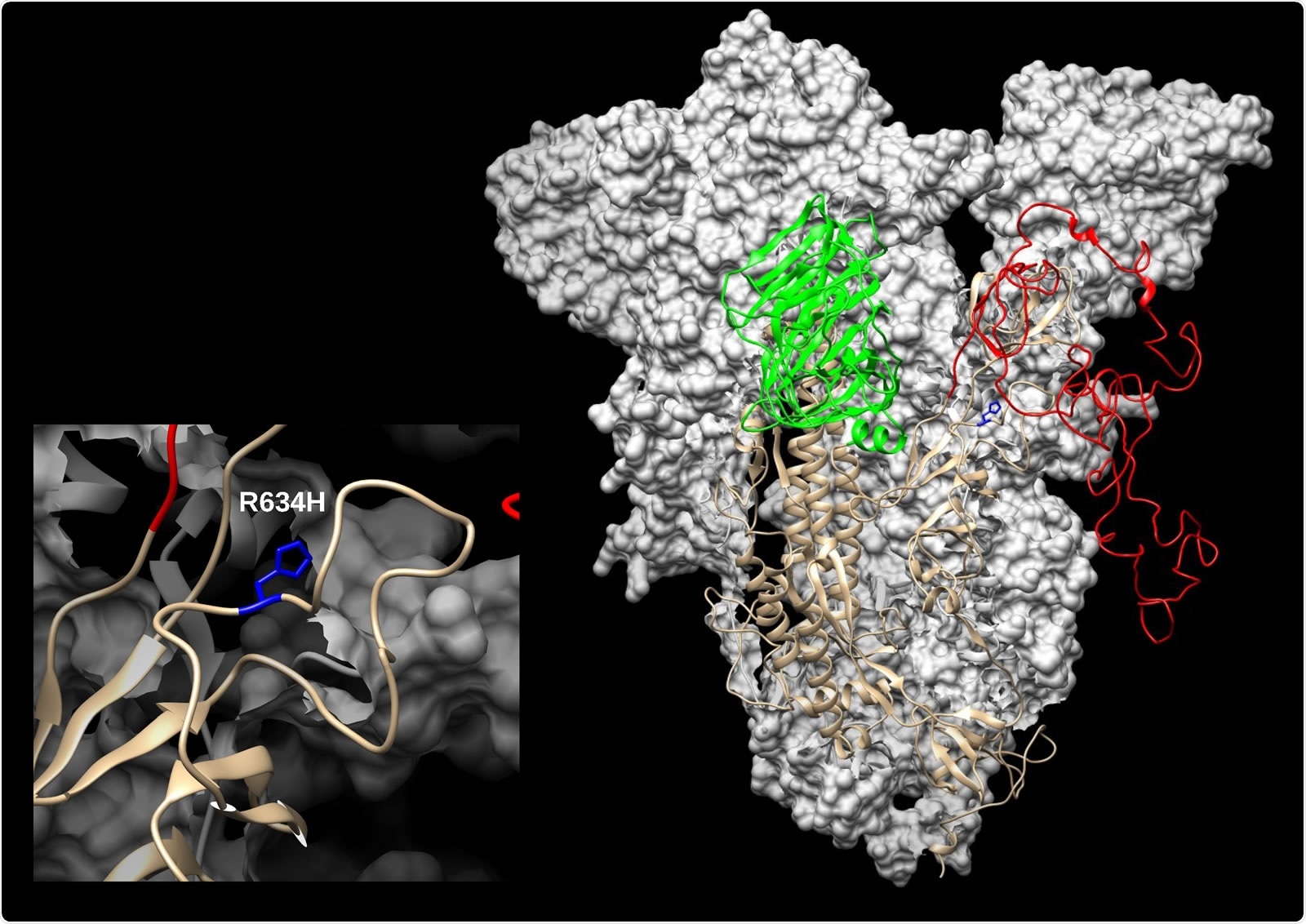
SARS-CoV-2 spike (S) modelled with SWISS-MODEL (Waterhouse et al., 2018) using 6ZGH structure as a template, drawn and colored in UCSF Chimera (Pettersen et al., 2004). N-terminal domain (NTD) is colored green and Receptor-binding domain/Cterminal domain (RBD/CTD) is red. The enlarged inset shows the location of R634H mutation (blue).

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Monitoring and analyzing SARS-CoV-2
Since the coronavirus disease 2019 (COVID-19) outbreak first began in Wuhan, China, late last year, it has spread at unprecedented rates and affected almost every country in the world.
The rapid viral spread and devastating health and socioeconomic consequences globally have led to urgent efforts to characterize the virus, track its spread and study its biological features to help identify potential treatment strategies and monitor the effectiveness of mitigation measures.
Checking SARS-CoV-2 genomes
In Sweden, by the end of April, the virus was reported to be infecting approximately 600 people per day and killing about 90 per day.
On 26th April, Soratto and the team collected and tested nasopharyngeal swabs from 23 patients with suspected COVID-19 at the Karolinska University Hospital in Stockholm, Sweden.
Seventeen of the patients were confirmed as positive for SARS-CoV-2 by reverse transcriptase-polymerase chain reaction (RT-PCR) testing. After extracting viral RNA and synthesizing complementary DNA, libraries with 350-base pair (bp) fragments were generated and prepared for shotgun sequencing.
After removing low-quality sequencers and adapters and the human genome reference, the researchers mapped the remaining reads to the complete genome of SARS-CoV-2 Whuan-Hu-1, as accessed in GenBank.
Once mapped, the Genome Detective Virus Tool was used to assemble the reads with the GenBank SARS-CoV-2 genome used as a reference, and single nucleotide variants were assigned to each genome.
The researchers were able to assemble four near-complete genomes that covered 99.7 to 99.8% of the GenBank reference genome and all of the coding region.
A variant was identified that could have destabilizing effects
Compared with the reference genome, the four assembled genomes each had between 9 and 14 nucleotide variants.
The team reports that all four genomes had mutations in the noncoding position 241 C (cytosine) to T (thymine) and three mutations in coding regions, which were two C to T at 3037bp and 14408bp, and one A (adenine) to G (guanine) at 23403bp.
In one of the genomes, a variant was found at 23463bp that was not found in any other GenBank SARS-CoV-2 genome. The mutation changes an arginine residue to a histidine residue at position 364 in the S1 subunit of the viral spike protein.
The researchers say this variant in the spike protein could have destabilizing effects.
“The variant is located at the surface of the S1 subunit, and could possibly affect the attachment of the virion to the cell, even though it does not change the receptor-binding domain itself,” writes the team.
Three genomes were strains previously thought to have declined in Sweden
On comparing the four SARS-CoV-2 genomes obtained from patients in Stockholm with those detected globally, the researchers found that they belonged to the genetic strains 20C/B.1/G and 20B/B.1.1/GR.
The Public Health Agency of Sweden had previously reported these genetic groups as two of the three main strains found in Sweden.
However, three of the genomes are from group B.1/G, which was also reported by the Public Health Agency of Sweden to have declined in prevalence by late April, say Soratto and team.
“This lineage has spread to more than 20 countries in Europe, the Americas, Asia, and Australia and corresponds to the Italian outbreak,” write the researchers.
“More investigation is needed in order to ensure that the spread of different types of SARS-CoV-2 is fully characterized,” they conclude.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Soratto T, et al. SARS-CoV-2 genome sequences from late April in Stockholm, Sweden reveal a novel mutation in the spike protein. bioRxiv 2020. doi: https://doi.org/10.1101/2020.08.03.233866
- Peer reviewed and published scientific report.
Soratto, Tatiany Aparecida Teixeira, Hamid Darban, Annelie Bjerkner, Maarten Coorens, Jan Albert, Tobias Allander, and Björn Andersson. 2020. “Four SARS-CoV-2 Genome Sequences from Late April in Stockholm, Sweden, Reveal a Rare Mutation in the Spike Protein.” Edited by Simon Roux. Microbiology Resource Announcements 9 (35). https://doi.org/10.1128/mra.00934-20. https://journals.asm.org/doi/10.1128/MRA.00934-20.