A new DNA methylation-based blood test can identify type 2 diabetes patients at the highest risk of heart attack or stroke years in advance, beating traditional risk scores and opening the door to earlier, more targeted prevention.
 Study: Epigenetic biomarkers predict macrovascular events in individuals with type 2 diabetes. Image Credit: Dragon Claws / Shutterstock
Study: Epigenetic biomarkers predict macrovascular events in individuals with type 2 diabetes. Image Credit: Dragon Claws / Shutterstock
In a recent study published in the journal Cell Reports Medicine, researchers identified epigenetic biomarkers predicting incident macrovascular events (iMEs) in people with type 2 diabetes (T2D).
People with T2D have two to four times higher cardiovascular disease (CVD) risk than non-diabetic individuals. Identifying individuals at risk for macrovascular events (MEs) is crucial for disease prevention. Nonetheless, ME prediction in people with T2D is suboptimal. While risk stratification scores for CVD are available, they have a moderate ability to stratify T2D patients. As such, there is a need to identify new biomarkers to improve CVD and ME prediction in T2D subjects.
The study and findings
In the present study, researchers identified blood-based epigenetic biomarkers to predict iMEs in people newly diagnosed with T2D. They included 752 participants with newly diagnosed T2D with available DNA methylation (DNAm) data and without known MEs from the All New Diabetics in Scania (ANDIS) and Uppsala County (ANDiU) cohorts in the “prospective cohort for MEs in T2D.” Of these, 102 individuals developed MEs over a mean follow-up of about four years and a maximum of seven years, while 650 did not.
The researchers analyzed DNAm of over 853,000 sites in blood to identify epigenetic biomarkers associated with future MEs. They found that DNAm of 461 sites was associated with iMEs using a Cox regression model adjusted for gender, age, body mass index (BMI), and glycated hemoglobin (HbA1c). These sites, annotated to 422 genes, were distributed across the genome.
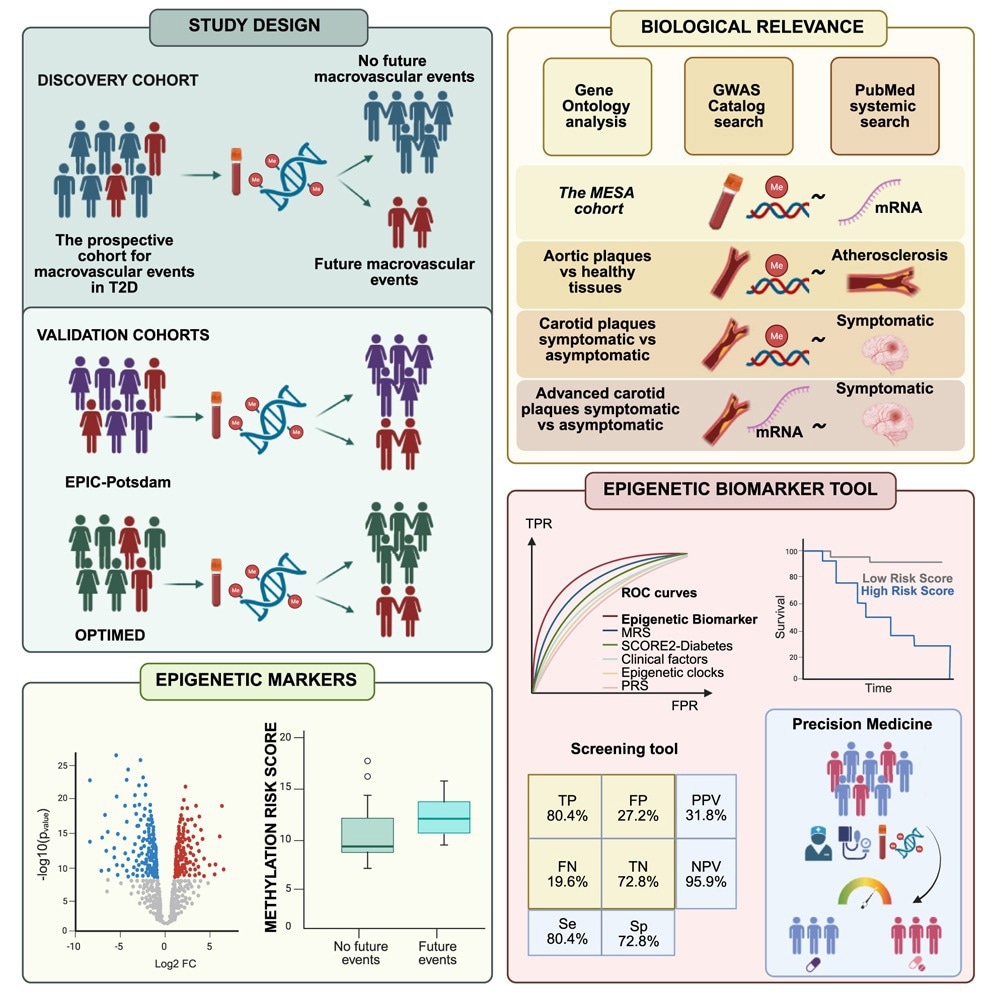
Garcı´a-Calzo´ n et al. identify a bloodbased epigenetic biomarker tool that helps predict future cardiovascular disease, which individuals with type 2 diabetes are more likely to develop. Their findings support the use of epigenetic biomarkers to improve risk stratification and guide more personalized prevention strategies in diabetes care.
Further, results remained consistent, with about 453 sites associated with iMEs, after additional adjustment for medications, smoking, lipid profiles, and cell composition. Next, the team examined whether a methylation risk score (MRS) would predict future MEs in individuals newly diagnosed with T2D. Methylation sites with absolute methylation differences ≥ 2% between controls and individuals with iMEs were filtered for inclusion in the MRS.
The MRS included 87 methylation sites; most sites (74%) were hypomethylated in those with iMEs relative to controls. Besides, the MRS was significantly different between controls and individuals with MEs. Next, the team performed five-fold cross-validation using logistic models to assess whether the MRS could differentiate between controls and individuals with iMEs in the prospective cohort for MEs in T2D.
For comparison, a cross-validation analysis was performed with only clinical risk factors of MEs. Moreover, the combination of clinical factors and MRS was examined. Receiver operating characteristic curves revealed that the MRS predicted iMEs with an area under the curve (AUC) of 0.81, while clinical risk factors alone predicted iMEs with an AUC of 0.69. The combination of the MRS and clinical risk factors yielded an AUC of 0.84.
Statistical testing showed the MRS performed significantly better than clinical risk factors alone (p = 0.001), and the combined model significantly outperformed clinical risk factors (p = 1.7 × 10⁻¹¹). Precision–recall analysis, appropriate for imbalanced data, also showed improved sensitivity and precision for MRS-based models.
Next, the ability of MRS to predict MEs was compared with established CVD risk scores, a polygenic risk score (PRS) based on 204 coronary artery disease-associated single-nucleotide polymorphisms (SNPs), and epigenetic clocks of mortality and aging. The United Kingdom Prospective Diabetes Study (UKPDS) and SCORE2-Diabetes risk scores that predict CVD in diabetic individuals had AUCs of 0.54 and 0.62, respectively, in predicting MEs.
The PRS, epigenetic clocks and mortality, and other risk scores (multi-ethnic study of atherosclerosis, atherosclerotic CVD, and Framingham risk scores) were significantly worse than MRS or MRS plus clinical factors, with AUCs ranging between 0.61 and 0.68. The optimal cutoff point for the combined biomarker tool, which showed the best prediction, was estimated to be 0.023, with a sensitivity and specificity of 0.804 and 0.728, respectively. At this cutoff, the model achieved a high negative predictive value of 95.9% and a moderate positive predictive value of 31.8%, meaning it could reliably identify individuals unlikely to experience iMEs but was less accurate for confirming those at risk. Net reclassification improvement analyses showed a 28.2% categorical and 90.2% continuous improvement over clinical risk factors alone. The test was estimated to cost approximately $200 per sample, a factor the authors suggest could be feasible for clinical screening if used selectively in high-risk populations.
Further, the researchers evaluated whether the 64 genes annotated to 87 methylation sites in MRS exhibit differential expression in carotid plaques from symptomatic and asymptomatic patients. They found differential expression of four genes in plaques from symptomatic patients compared to asymptomatic patients. They also reported that 72% of the MRS genes had prior links to CVD in literature or GWAS data, and that several methylation sites overlapped with those differentially methylated in aortic plaque tissue.
Next, the team performed validation analyses of the methylation sites associated with iMEs in OPTIMED and EPIC-Potsdam cohorts. They validated 43 and 32 methylation sites in OPTIMED and EPIC-Potsdam cohorts, respectively, using Cox regression models adjusted for BMI, age, gender, and HbA1c. Moreover, five out of 87 sites in the MRS were associated with iMEs in the OPTIMED cohort.
An MRS developed using these five sites (MRS5sites) was significantly different between controls and individuals with iMEs in the OPTIMED cohort and the prospective cohort for MEs in T2D. The combination of clinical risk factors and MRS5sites had an AUC of 0.8 in the OPTIMED cohort and 0.78 in the prospective cohort of MEs in T2D. Notably, OPTIMED included newly diagnosed T2D participants, whereas EPIC-Potsdam was a general population cohort, supporting the broader applicability of the findings.
Conclusions
In sum, the study identified and validated a blood-based epigenetic biomarker predicting the risk of the first ME, in combination with and independently of clinical risk factors, in individuals newly diagnosed with T2D. T2D subjects with an MRS in combination with clinical risk factors of more than 0.023 were likely to develop MEs over the maximum follow-up of seven years. Overall, the epigenetic biomarker predicts iMEs better than established risk scores, supporting its future clinical use.
The authors note, however, that external validation in other ethnicities is needed, and that the moderate PPV likely reflects the low prevalence of events in this newly diagnosed population. They also caution that environmental factors such as diet, physical activity, and medications can influence DNA methylation, and that these were not fully captured in the current cohorts.