A team of scientists at the University of California, Berkeley, Innovative Genomics Institute, Illumina, Inc., and Stanford University has performed a genomic sequencing of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) collected from sewage in the San Francisco Bay Area. The study findings indicate that the genotypes detected in the sewage are identical to the genotypes detected in clinical samples obtained from that region. The study is currently available on the medRxiv* preprint server.
Genomic sequencing of SARS-CoV-2 RNA is an important approach for determining viral evolution and transmission dynamics. At the initial period of the coronavirus disease 2019 (COVID-19) pandemic in the United States, the whole genome sequencing data of SARS-CoV-2 has revealed that there are multiple viral introduction events in the New York City and California. Also, the frequency of community transmission is higher for a particular type of viral genotype after its first introduction in Washington state. Moreover, distinct SARS-CoV-2 strains have been identified in a single neighborhood in San Francisco.
Given the presence of SARS-CoV-2 RNA in human feces, estimation of the viral abundance in many municipal regions has been undertaken through quantitative reverse transcriptase-polymerase chain reaction (RT-qPCR)-based wastewater sequencing.
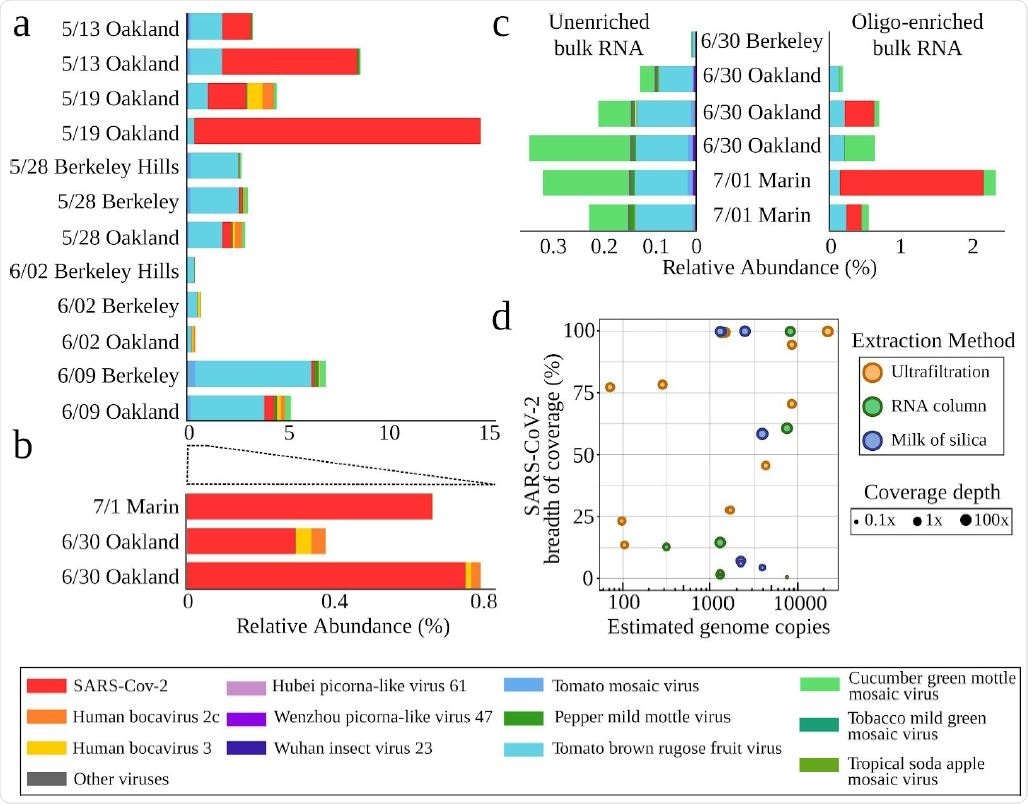
Characterized viruses detected in enriched and unenriched wastewater metatranscriptomes. Relative abundances of viruses with eukaryotic hosts in the RefSeq database as a percentage of total sequencing reads derived from the sample in ( a) amicon ultrafiltration (viral fractionation) and ( b) total RNA column and milk of silica samples. All samples were enriched with the Illumina Respiratory Virus Panel. ( c) Relative abundances of RefSeq viruses in unenriched metatranscriptomics (left) and the same samples after oligo-enrichment with the Illumina Respiratory Virus Panel. (d) The relationship between the quantity of viral genome copies in 40 μL of purified RNA and SARS-CoV-2 genome completeness (measured in breadth of coverage) for each sample. Samples are colored by extraction methodology, and the size of the point corresponds to the mean SARS-CoV-2 depth of coverage.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Current study design
To identify viral abundance and genotype prevalence in various regions in the San Francisco Bay Area, the current study performed genomic sequencing of SARS-CoV-2 RNA isolated directly from wastewater. The scientists extracted viral concentrates and total RNA from composite samples of raw sewage collected between May 2020 and July 2020.
Important observations
They utilized metagenomic sequencing approaches to detect viral abundance and single nucleotide variants, and the results revealed that SARS-CoV-2 was present at varying abundances (0% - 14%) in study samples.
Besides SARS-CoV-2, other human and plant viruses were identified in wastewater samples, including human bocaviruses, picorna-like viruses, cucumber green mottle mosaic virus, and pepper mild mottle virus.
In about 31% of samples, the scientists were able to obtain complete consensus SARS-CoV-2 genomes (covered length >99%). About 4 base pair differences were observed between consensus genomes obtained from Alameda County and Marin county. This indicates that consensus genomes may represent the predominant lineages of SARS-CoV-2 in the study population during summer season of 2020.
To identify alternative viral genotypes, the scientists utilized a pipeline for metagenomic single nucleotide variant calling and identified different genomic variants of SARS-CoV-2 in each wastewater sample. These variants were identical to those obtained from clinical patient samples. About 50%, 61%, and 71% of variants detected in wastewater samples were also detected in clinical viral genomes obtained from California, the United States, as well as those collected worldwide, respectively. This indicates that the actual genomic variation of SARS-CoV-2 in a given population can be summarized from wastewater viral genome sequencing data.
The scientists also used a hypergeometric test to analyze the probability of overlap by chance between wastewater genomic variants and clinical genomic variants in a given region. They observed that the probability of non-random overlap was highest in Alameda county.
Interestingly, the scientists observed that besides genomic variants of SARS-CoV-2 that were identical in both wastewater and clinical patient samples, there were four recurrent variants in wastewater samples that were not detected clinically in California. However, these variants are previously identified in other regions in the United States and seem to have arrived in those regions only in July 2020. This indicates that these variants may be detected clinically in California in the future.
Study significance
The current study reveals that different genotypes of SARS-CoV-2 strains can be identified in wastewater samples. Wastewater sequencing can even reveal genetic variants that are still undetected in clinical patient samples.
Because the wastewater sequencing method does not depend on specific polymerase chain reaction primers, the failure of detecting viral strains with mutations can be avoided. Moreover, this approach can provide important information about viral distribution within a given population, as well as can reveal recent entry of different SARS-CoV-2 genotypes in a given region.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources