More than 47.3 million people around the world have been infected with the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that causes COVID-19. The virus has claimed over 1.2 million lives, and the pandemic shows no signs of abating at present. Many countries around the world that were on the path to recovery with declining numbers of cases are now witnessing a second wave of infections that have prompted worldwide lockdowns once again to break the chain of transmission.
A team of researchers working with Scott Wesley Long from Houston Methodist Hospital and colleagues from Texas, New York, and Illinois studied the patterns of genetic mutations of SARS CoV-2 and its impact on the second wave of infections worldwide. Their study titled, "Molecular Architecture of Early Dissemination and Massive Second Wave of the SARS-CoV-2 Virus in a Major Metropolitan Area," was released in the latest issue of the journal mBio.
Background
SARS-CoV-2 is highly infectious and spreads rapidly from person to person. In the United States (US), viral spread hot spots have been in Seattle, the New York City (NYC) regions and others, leading to thousands of infections and deaths nationwide. The research team wrote that by mid-August there were "227,419 confirmed SARS-CoV-2 cases in NYC, causing 56,831 hospitalizations and 19,005 confirmed fatalities and 4,638 probable fatalities". In Seattle and King County there have been, "17,989 positive patients and 696 deaths".
Travel and COVID-19
According to the researchers, the primary cases have been seen among those who had been associated with national or international travel. This, they wrote, has led to the outbreaks.
Genomic variations
The Houston area in Texas is one of the US's most ethnically diverse regions and has witnessed several thousands of cases. One of the central molecular diagnostic laboratories has been catering to the Houston Methodist hospitals, and data could be obtained from that lab. The genomic variations causing the infections in the region could be determined, wrote the researchers.
Genome sequencing was done to discern variations in the strains of the virus causing infections around the region. The genomic sequencing studies began in March and by May, it had to be expanded when the researchers realized that there was a chance of a second wave after the first wave of infections was declining.
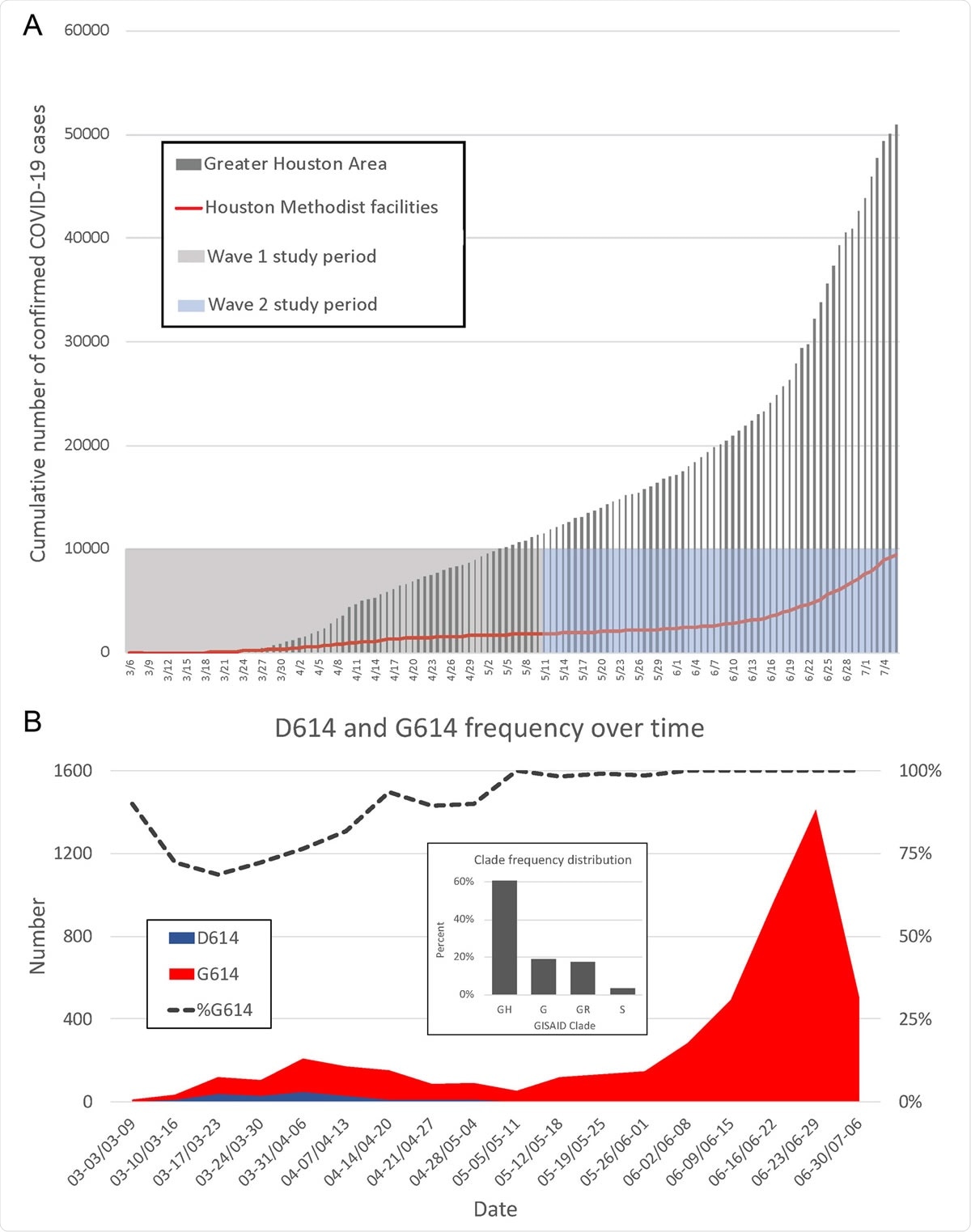
(A) Confirmed COVID-19 cases in the Greater Houston Metropolitan region. Data represent cumulative number of COVID-19 patients over time through 7 July 2020. Counties include Austin, Brazoria, Chambers, Fort Bend, Galveston, Harris, Liberty, Montgomery, and Waller. The shaded area represents the time period (indicated as month/day along the x axis) during which virus genomes characterized in this study were recovered from COVID-19 patients. The red line represents the number of COVID-19 patients diagnosed in the Houston Methodist Hospital Molecular Diagnostic Laboratory. (B) Distribution of strains with either the Asp614 or Gly614 amino acid variant in spike protein among the two waves of COVID-19 patients diagnosed in the Houston Methodist Hospital Molecular Diagnostic Laboratory. The large inset shows major clade frequency for the time frame studied (indicated as month-day to month-day along the x axis).
Study objective
This study reported that in the Houston region alone, different strains of the virus were introduced again and again from outside. They could determine this from the genotypes that matched those of the virus strains present in genetic clades in Europe, Asia, South America and elsewhere in the US.
Specific mutating strains
Strains detected in Houston had a Gly614 amino acid replacement in the spike protein, the researchers found. This caused increased the infectivity or contagiousness of the virus, spreading rapidly from person to person and giving rise to the second wave.
Patients detected with Gly614 variant strains were found to have higher viral loads in their noses, nasopharynx when diagnosed.
What was done?
This study involved a detailed genotyping of 5,085 SARS-CoV-2 strains that led to the two infection waves of COVID-19 disease in metropolitan Houston, Texas. The ethical diversity led to the choice of this region for this study. There is a population of seven million residents in this region, wrote the researchers.
What was found?
Results revealed that viruses with distinct genotypes were introduced into the Houston population independently and more than once. This could have been due to travel. Overall results were:
- Wave 1 is defined as new cases between 5th March and 11th May 2020. Wave 2 is defined as cases between 12th May and 7th July 2020.
- The strains detected in the second wave were more likely to carry the Gly614 amino acid replacement in the spike protein
- The Gly614 polymorphism (the D614G mutation) was found to raise the risk of transmission and infectivity of the virus
- Those with the variant virus also had higher viral loads in their nasopharynx when they were diagnosed initially
- The altered virus, however, was not found to be more virulent than the earlier strain
- Disease severity among those infected in the second wave was more due to underlying disease conditions or the genetic makeup of the individual infected than the genetic makeup of the virus itself.
- The researchers also noted that the regions of the gene that coded for the spike protein were most amenable to mutations. This could be of vital importance because these proteins are the ones that the vaccines against COVID-19 are possibly targeting.
Conclusions and implications
A co-author of the study Ilya Finkelstein, Associate Professor of Molecular Biosciences at The University of Texas at Austin, said that this study reveals that "the virus is mutating due to a combination of neutral drift - which just means random genetic changes that don't help or hurt the virus - and pressure from our immune systems."
Finkelstein also said that "The virus continues to mutate as it rips through the world... Real-time surveillance efforts like our study will ensure that global vaccines and therapeutics are always one step ahead." A lead author James Musser of Houston Methodist explained in a statement: "We have given this virus a lot of chances. There is a huge population size out there right now."
Long and his fellow researchers concluded: "Our study is the first analysis of the molecular architecture of SARS-CoV-2 in two infection waves in a major metropolitan region. The findings will help us to understand the origin, composition, and trajectory of future infection waves, and the potential effect of the host immune response and therapeutic maneuvers on SARS-CoV-2 evolution."