Over the past two decades, three major betacoronavirus outbreaks have occurred: the severe acute respiratory syndrome (SARS) in 2002, the Middle East respiratory syndrome (MERS) in 2012, and the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in late 2019. SARS-CoV-2, the pathogen that causes coronavirus disease 2019 (COVID-19), spread rapidly across the globe by early 2020; the World Health Organization (WHO) declared the outbreak a global pandemic in March 2020. Nearly a year on since it was first detected in humans in Wuhan, China, SARS-CoV-2 has infected over 63.93 million people across the globe and has killed over 1.48 million.
A team of researchers at the University of Tokyo in Japan have detected a sarbecovirus, called Rc-o319, which is phylogenetically positioned in the same clade as SARS-CoV-2 in Rhinolophus cornutus bats in Japan.
Study background
Betacoronaviruses are a large family of pathogens characterized by their lipid envelopes, positive-strand RNA, and spike proteins. Two viruses of the subgenus SArbecovirus are SARS and SARS-CoV-2.
The Rhinolophus bats or horseshoe bats are natural reservoirs of sarbecoviruses in Asia, Europe, and Africa, but their epidemiology and distribution remain unknown.
R. cornutus is an endemic bat species in Japan; they inhabit caves and abandoned tunnels during daytime and hunt insects at night.
The study, which was published in the journal Emerging Infectious Diseases, reported that they captured four R. cornutus bats in a cave and obtained RNA samples from fresh feces. They utilized a real-time reverse transcription PCR (rRT-PCR) to detect the gene of sarbecovirus in 2013.
In 2020, the team performed RNA sequencing and found the full genome sequence of one sample, Rc-o139. They performed a BLAST analysis on the entire genome of Rc-o139, which showed the highest nucleotide homology to the SARS-CoV-2 HKG/HKU strain. The data gathered thus showed that Rc-o319 is genetically related to SARS-CoV-2.
Identifying the pathogen origin
The team emphasized the importance of knowing the origin of pathogens, mainly due to recurrent breakouts of betacoronaviruses over the past twenty years.
“Identifying the origins of an emerging pathogen can be critical during the early stages of an outbreak, because it may allow for containment measures to be precisely targeted at a stage when the number of daily new infections is still low,” the team noted in the study.
Locating the precise origin of a virus can help mitigate the spread of the outbreak and, at the same time, help prevent future outbreaks.
In previous outbreaks, early detection through genomics was not possible, such as the SARS outbreak in 2002 and the avian influenza H5N1 outbreak in 1997. In 2009, however, during the AH1N1 outbreak, rapid genomic analysis had become a part of outbreak response.
The study showed that viruses closely related to SARS-CoV-2 have been circulating in horseshoe bats for many years. These findings also corroborate a major new discovery early in November 2020 of a closely related betacoronavirus to SARS-CoV-2 found in a frozen bat sample collected in 2010 in Cambodia.
“The existing diversity and dynamic process of recombination amongst lineages in the bat reservoir demonstrate how difficult it will be to identify viruses with potential to cause major human outbreaks before they emerge,” the researchers added.
The current study underlines the need for a global network of real-time human disease surveillance systems. These could help detect outbreaks before they get out of hand. Further, these surveillance systems can rapidly deploy genomic tools and functional studies to identify and characterize viruses.
The coronavirus pandemic is an eye-opener on what is needed in reducing the risk of future outbreaks.
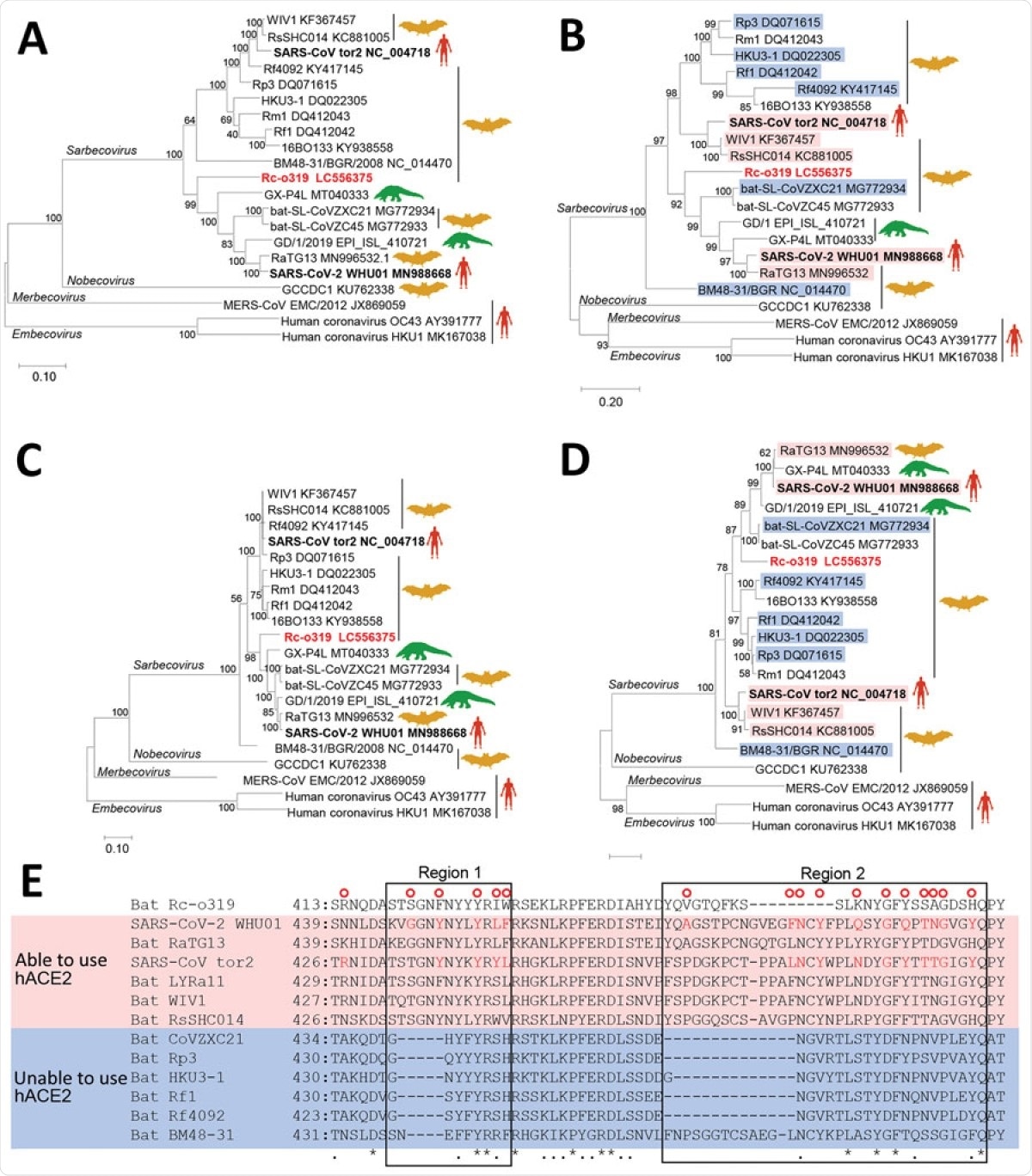
Phylogenetic analysis of sarbecovirus sequenced from little Japanese horseshoe bats (Rhinolophus cornutus) and genetically related to human SARS-CoV-2, Japan. A–D) Phylogenetic trees were generated by using maximum-likelihood analysis combined with 500 bootstrap replicates and show relationships between bat-, human-, and pangolin-derived sarbecoviruses. Phylogenetic trees are shown for nucleotide sequences of the full genome (A), the S protein gene and amino acid sequences (B), the ORF1ab (C), and the S protein (D). Red text indicates positions of Rc-o319, the sarbecovirus sequenced in this study. For panels B and D, magenta bands indicate viruses with S proteins that bind to human ACE2; blue bands indicate viruses with S proteins that do not bind to human ACE2. Bootstrap values are shown above and to the left of the major nodes. Scale bars indicate nucleotide or amino acid substitutions per site. E) Amino acid sequence alignment of the RBM of S proteins that are able or unable to bind to human ACE2. Amino acid residues of the RBM that contact human ACE2 of SARS-CoV-2 and SARS-CoV are indicated in the upper side by red circles. The 2 regions of S protein RBM known to interact with human ACE2 are indicated by boxes labeled region 1 and region 2. ACE2, angiotensin-converting enzyme 2; hACE2, human angiotensin-converting enzyme 2; ORF1ab, open reading frame 1ab; RBM, receptor-binding motif; S, spike protein; SARS-CoV, severe acute respiratory syndrome coronavirus; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
The global toll
Despite how advanced healthcare systems and laboratories are today, the COVID-19 pandemic has shown us that they are still not ready for a health crisis of this magnitude. Further studies and investigations are crucial to improve the response of governments across the globe to the current pandemic and ones that may arise in the future.
The United States, one of the most developed countries worldwide, reports the highest number of infections, reaching 13.72 million cases. The virus has claimed more than 271,000 lives in the country.
India, Brazil, and Russia report surging cases, topping 9.46 million, 6.33 million, and 2.30 million cases, respectively.
Mexico reports a high death rate due to COVID-19. Of the 1.11 million confirmed cases, the death toll has reached more than 105,000.
Journal reference:
- Murakami, S., Kitamura, T., Suzuki, J., Sato, R., and Aoi, T. (2020). Detection and Characterization of Bat Sarbecovirus Phylogenetically Related to SARS-CoV-2, Japan. Emerging Infectious Diseases. https://dx.doi.org/10.3201/eid2612.203386,
https://wwwnc.cdc.gov/eid/article/26/12/20-3386_article