With second and third waves of the COVID-19 pandemic hitting many shores, the possibility that severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) will become endemic has become much more likely. Moreover, scientists are now looking ahead to future lethal coronavirus epidemics. This has intensified interest in the fundamental structural biology of the virus.
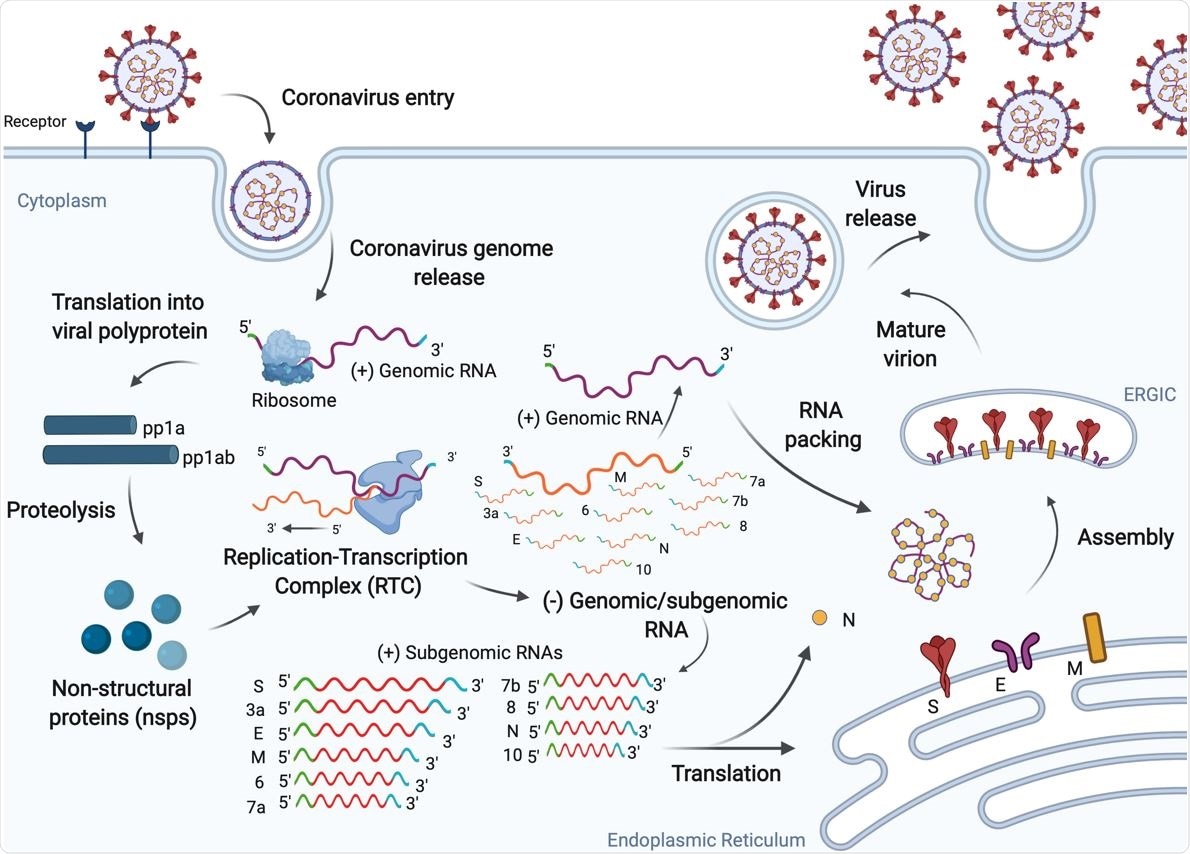
SARS-CoV-2 replication cycle. The virus enters human cells via endocytosis by binding the ACE2 receptor and releasing its positive-sense RNA genome. The virus exploits the host machinery to facilitate efficient viral replication, which ultimately leads to progression of infection.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Limited Therapeutics for COVID-19
There are only a very few drugs that have received even emergency use approval by the U.S. Food and Drugs Administration (FDA) for use in COVID-19, notwithstanding the widespread off-label use of many old and new drugs. One of the approved drugs is remdesivir, an inhibitor of RNA-dependent RNA polymerase (RdRp), originally developed to counter Ebola. The efficacy of this drug is moderate at best, with no evidence that it reduces mortality. However, further studies are required to decide on this aspect of the drug.
Another important aspect, besides efficacy, is the scalability and ease of manufacturing of a drug. Remdesivir requires multiple steps and a stereospecific synthetic process. Small molecules are now entering the field, such as baricitinib (Olumiant), which inhibits the inflammatory cascade associated with disease severity. It is now being repurposed from its original role in the treatment of rheumatoid arthritis, to serve as a companion drug to remdesivir in COVID-19.
The difficulty of finding effective antivirals in the current pandemic has stimulated the search for better drug development approaches that will reveal targetable viral structures, thus helping to prevent and treat both the current and any future coronavirus outbreaks.
Earlier, viral protein targets have been identified, such as viral endocytosis preceding its entry into the host cytosol, viral protein assembly; and viral genome condensation for virion assembly.
Small Molecules with RNA Targets
The current study shifts to small molecules that target RNA. Small molecules that behave like drugs can be used to explore the function of various proteins and large molecules, acting as chemical probes. They also offer a scaffold on which to build a clinically effective and safe drug with high bioavailability.
Small molecules that are directed against RNA are slowly making their appearance as therapeutics, with one drug targeting non-ribosomal RNA approved a few months ago to help patients with spinal muscular atrophy (SMA). A host of viral RNAs and other RNAs involved in disease processes have also been demonstrated to be targeted effectively by small molecules.
Small Molecule Inhibiting 5’ UTR-Mediated Translation
The researchers aim to exploit the conservation of certain SARS-CoV-2 RNA motif sequences and structures. Specifically, they identified the presence of stem-loops at the 5’ end of the RNA genome and other structures involved in frameshifting and replication.
Four of the five stem-loops of the 5’ UTR have 70% to 100% conserved sequences across the various members of the genus. Their position in the 5’ UTR suggests they contain homologous nucleotides with functional equivalency. These are apparently cis-elements involved in regulating viral replication. Such regulation may occur through protein-RNA interactions or allosteric RNA-RNA effects between stem-loops on the 5’-UTR.
Very recently, studies have shown the importance of the 5’-UTR, in that its presence in the translation pathway within the host cell multiplies the protein yield fivefold. This underlines its pivotal role in the effective translation of viral proteins and viral takeover of the host cell translational processes.
In particular, the 5’ UTR has several targetable structures, which can be bound by small molecules, resulting in antiviral activity against several RNA viruses like the hepatitis C virus and EV71. Other viral RNAs that can be effectively targeted include the frameshifting elements (FSE) of SARS-CoV-2. For instance, a small molecule engaging the attenuator hairpin structure near the pseudoknot of the FSE can be used in combination with ribonuclease targeting chimera (RIBOTAC) technology. This will
Amiloride Analogs
Amiloride is a common RNA-binding scaffold and has been synthetically refined to target the enterovirus EV71. The researchers used dimethylamiloride (DMA)-based antivirals that would target the RNA at the internal ribosomal entry site (IRES) region in the 5’-UTR of the enterovirus EV71. This is an approach that allows synthetic tuning of a known RNA construct.
The DMA base was further functionalized to provide a bioactive antiviral compound, which formed a complex with viral RNA at IRES stem-loop II, and with the host protein, AUF1. This ternary complex represses translation and thus prevents viral replication.
Reduction of Viral Titer
A preliminary focused DMA library screening, using OC43-infected cells, led to the identification of three lead compounds, namely DMA-132, -135, and -155, that reduced viral titer when tested against the human seasonal betacoronavirus OC43. This was selected for its low virulence, obviating the requirement for biosafety level 3 facilities.
The essential elements in the structure-activity relationships were found to involve the C5 dimethylamine group and C6 rigid aromatic substituents.
These analogs were confirmed to reduce the titer of SARS-CoV-2 by 10-30 times in a dose-dependent manner in a cell assay. The half-maximal inhibitory concentration (IC50) was 10 and 15 micromoles for DMA-135 and 155, respectively. At these doses, cytotoxicity was absent, indicating their potential for therapeutic use.
In a reporter gene experiment, DMA addition resulted in a significant reduction in viral gene translation by 50% to 90%. Another important finding is that this translational repression by small molecules depends only on the presence of the 5’ end of the viral sequence.
5’ UTR-Binding Vital to Antiviral Activity
The antiviral activity was shown to be a function of the 5’-UTR and the proximal region of the genome. Further exploration reveals targetable binding sites in bulge-like regions in stem-loops 4 and 5a in the 5’-UTR, and in stem-loop 6 of ORF1a.
The DMA-stem loop interactions were examined by NMR, showing that the DMAs caused chemical shift disruptions for each of the stem-loops, occurring via specific surfaces of the 5’ region. DMA binding occurs preferentially to large internal or bulge loops in the stem-loops. The degree to which such perturbations occur also varies, with some DMAs causing different surfaces to interact and others leading to a broadening of the signals. The most significant interaction was in stem-loop 6, which has a 16-nucleotide bulge loop, showing weak pairing. The 5’ end of this loop may interact with the nucleocapsid antigen and thus regulate the genome's packing to form intact virions.
They also carried out a computational docking analysis which supports the NMR data and suggests that the three DMAs bind to three separate but nearby pockets. The stem-loops with predicted bulges were found to be most suitable for drug interaction. The three candidate compounds with the highest antiviral activity had the highest score when examined against the stem-loops with the largest chemical shift perturbations following DMA binding. This further corroborates the potential fitness of these analogs as targeting ligands for SARS-CoV-2 stem-loop RNA.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources