Kirsten St. George, Nathan D. Grubaugh and colleagues say that regardless of increased travel restrictions and increased coronavirus testing, the United States underreported genomic sequencing of potential new variants from confirmed positive COVID-19 cases. Because the United States sequenced only approximately 0.13% of COVID-19 cases, it was initially difficult to detect when the B.1.1.7 variant first emerged in America.
“The sequencing capacity is highly variable across the country, and our travel data helps to identify regions disproportionately underreporting cases of B.1.1.7 and where it would be prudent to immediately prioritize variant surveillance,” write the authors.
The study “Early introductions and community transmission of SARS-CoV-2 variant B.1.1.7 in the United States” is available as a preprint on the medRxiv* server, while the article undergoes peer review.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Tracking down the virus
The researchers used a combination of October 2020 flight records from the United Kingdom into all United States airports and SARS-CoV-2 genomic sequencing to detect the origins of the B.1.1.7 variant in the U.S. They reasoned this time period was chosen because not only was it the most recent data available, but it also overlapped with the time the B.1.1.7 variant was initially thought to have been spreading all over the United Kingdom.
According to the CDC’s early February report, 33 states have confirmed 541 B.1.1.7 cases in the United States. However, the authors reason the low genomic surveillance may not represent the true extent of the variant’s spread. They instead analyzed the number of reported cases from December 2020 to January 2021 with the percent of SARS-CoV-2 clinical samples taken.
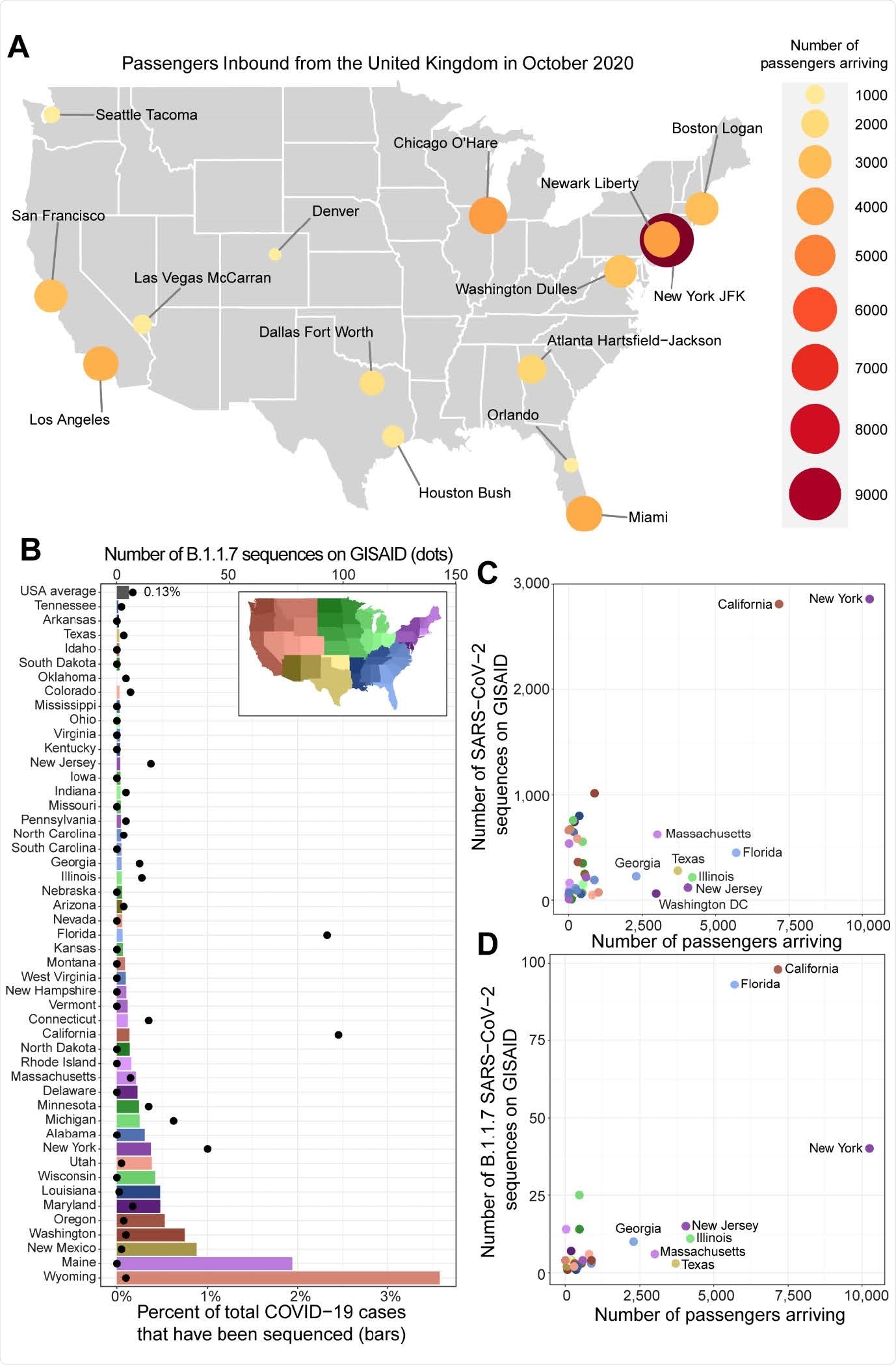
Identification of regions at risk for importation of B.1.1.7 and at risk for underreporting B.1.1.7. A. Number of passengers inbound from the United Kingdom for the top 15 airports in October 2020 (labeled dots; size and color scaling with population, see legend) are shown on a map of the continental United States (grey). B. Bar plot represents the percentage of cases in each state during Dec 2020 and Jan 2021 (bottom x-axis; sourced from https://covidtracking.com/data) that have sequences uploaded to GISAID.org (accessed on Feb 4, 2021). Bars are colored according to region (legend, top right). The number of B.1.1.7 sequences for each state (top x-axis; black dots) was determined by the Pangolin lineage assignment in the GSIAID.org metadata. C-D. Total number of passengers arriving from the UK in October 2020 to each state in the continental US is plotted against the number of SARS-CoV-2 (C) or B.1.1.7 SARS-CoV-2 (D) sequences available on GISAID.org. Points are colored according to region (legend from panel B). The data used to create this figure are listed in Data S1.
Strong evidence of variant spread first originating in New York
Low genomic surveillance was reported from December to January, with only 0.1% of COVID-19 cases sequenced and made available to researchers. A total of 26 states had less than 0.1% case sequences, and at the time of preprint publication, 14 states had not submitted any genomic sequence related to B.1.1.7.
A high number of B.1.1.7 strains were found in New York, California, and Florida. The sequencing coincides with the large number of incoming international flights from the United Kingdom into these states’ airports.
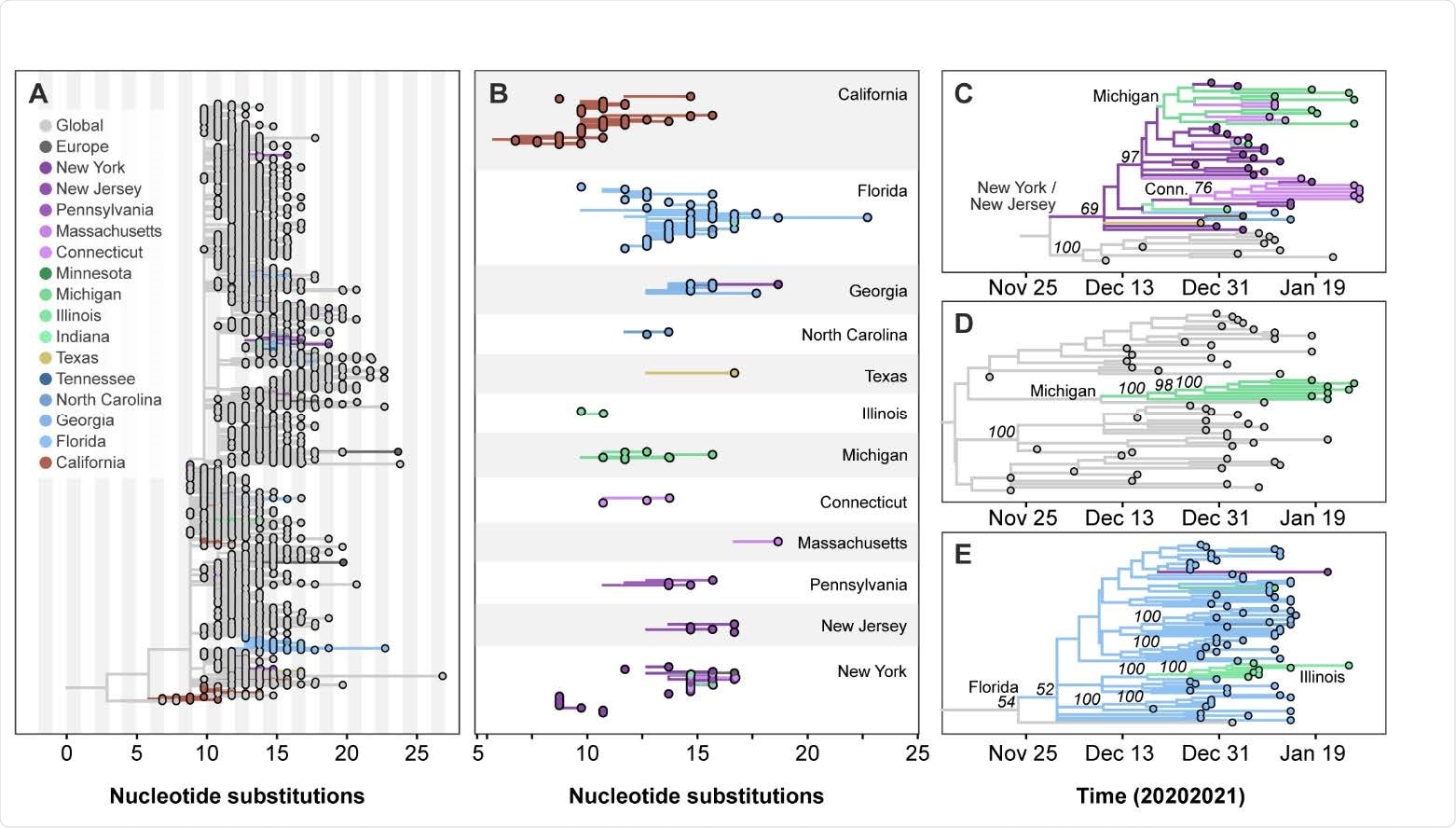
Multiple introductions, domestic spread, and community transmission of B.1.1.7 SARS-CoV-2 in the United States. A. Maximum likelihood phylogeny of B.1.1.7, including representative genomes from the US, Europe, other global locations. B. Highlights of singletons and clades representing direct introductions of B.1.1.7 from Europe, into distinct regions of the US, based on the same phylogenetic tree shown in (A). A list of the Europe to US transitions can be found in Data S1. C-E. Time-informed maximum likelihood phylogeny of distinct B.1.1.7 clades showing instances of domestic spread (C,E) and/or community transmission within New York (C), Connecticut (C), Michigan (C,D), and Illinois (E). The list of SARS-CoV-2 sequences used in this study and author acknowledgments can be found in Data S2.
Of all three, the researchers concluded that New York most likely was the central hub for incoming B.1.1.7 variants, which later spread to other states. The New York-New Jersey area had a record of 14,317 passengers to and from the United Kingdom. These results correspond with the first COVID-19 case with the new variant detected in New York’s Saratoga County on December 24, 2020.
As the B.1.1.7 variant remained undetected, the researchers suggest sustained community transmission likely occurred in New York, New Jersey, Connecticut, and Illinois starting January 2021.
“Thus, increasing surveillance for B.1.1.7 and other variants through sequencing and more conventional methods must be made a high priority,” write the researchers.
Strong possibility of B.1.17 variant in other states
While the researchers sequenced most B.1.1.7 variants in New York, the researchers say underreporting may have contributed to undetected cases in other states. Their results showed Texas, Illinois, and New Jersey had more than 2,500 people flying in from the United Kingdom. Yet, all three states had 0.05% or less of sequenced cases. The authors suggest a strong possibility that the spread of the new strain is increasing in other states.
“In places like New Jersey, Illinois, and Texas, however, if SARS-CoV-2 genomic surveillance could be increased, it might determine if B.1.1.7 cases are disproportionately underreported as compared to New York and California,” write the researchers.
More genomic surveillance needed for future research
Given the limited travel data and low genomic surveillance, the researchers say it’s too small of a sample to represent the entire spread of this variant in the United States. Additionally, they could not evaluate the rate of transmission across different regions and assumed community spread was high. “In reality, local conditions and behaviors play an important role for B.1.1.7 establishment, and could explain why cases are low in some states.”
With the rising spread of the new variants, the research team suggests increasing genomic surveillance is a high-priority task needed to mitigate the virus’s spread.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources