According to research from the University of California, some cases of severe coronavirus disease 2019 (COVID-19) may be related to the immunological memory of previous H3N2 influenza A infection.
The team says this “original antigenic sin” may explain the diverse disease outcomes that have been observed following infection with the causative agent severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
Gregory Weiss and colleagues say the study indicated that the αEp9 antibodies that are observed in severe COVID-19 might have originated from previous infection with H3N2 influenza A.
The researchers suggest that future studies investigate the correlation between a country’s rate of the H3N2 2014 influenza and severe COVID-19.
Examining epitope conservation and antibody cross-reactivity could enable the prediction of OAS-based immune responses and disease outcomes.
Identifying detrimental, benign or beneficial OAS pathways could also be useful in guiding vaccine design, adds the team.
A pre-print version of the research paper is available on the bioRxiv* server, while the article undergoes peer review.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
More about original antigenic sin
Original antigenic sin (OAS) refers to when immune responses that were previously adapted to combat a primary or “original” infection then later target a similar, but not identical, microbe.
Since B-cells undergo affinity maturation following a primary infection, affinity-matured antibodies from previous infections can sometimes out-compete the naïve antibodies that are generated to target a new antigen specifically.
“The phenomenon has been observed for immune responses to dengue fever, influenza, respiratory syncytial virus and human immunodeficiency virus,” says Weiss and colleagues.
Ideally, OAS would expedite pathogen clearance by targeting highly conserved antigens, but suboptimal targeting by non-neutralizing antibodies binding can exacerbate disease, either by enhancing viral infection or by over activating the innate immune response.
“The wide range of outcomes observed in SARS-CoV-2 infected individuals, from asymptomatic to fatal, has been hypothesized to result from a patient’s unique immunological memory,” says the team.
Recently, the researchers demonstrated a strong association between severe COVID-19 and the presence of antibodies that bind to an epitope region of the SARS-CoV-2 nucleocapsid protein called Ep9.
“These two observations suggest an OAS-based mechanism for the disease severity observed in αEp9(+) patients,” writes Weiss and colleagues.
What did the researchers do?
The researchers looked for homologs to Ep9 in protein sequence and structural homology databases, revealing candidate OAS epitopes.
The potential OAS epitopes to αEp9 Abs were tested by phage ELISA of pooled plasma from three sets of five αEp9(+) and five αEp9(-) COVID-19 patients.
The team identified a putative original antigen that was capable of stimulating the production of cross-reactive αEp9 antibodies.
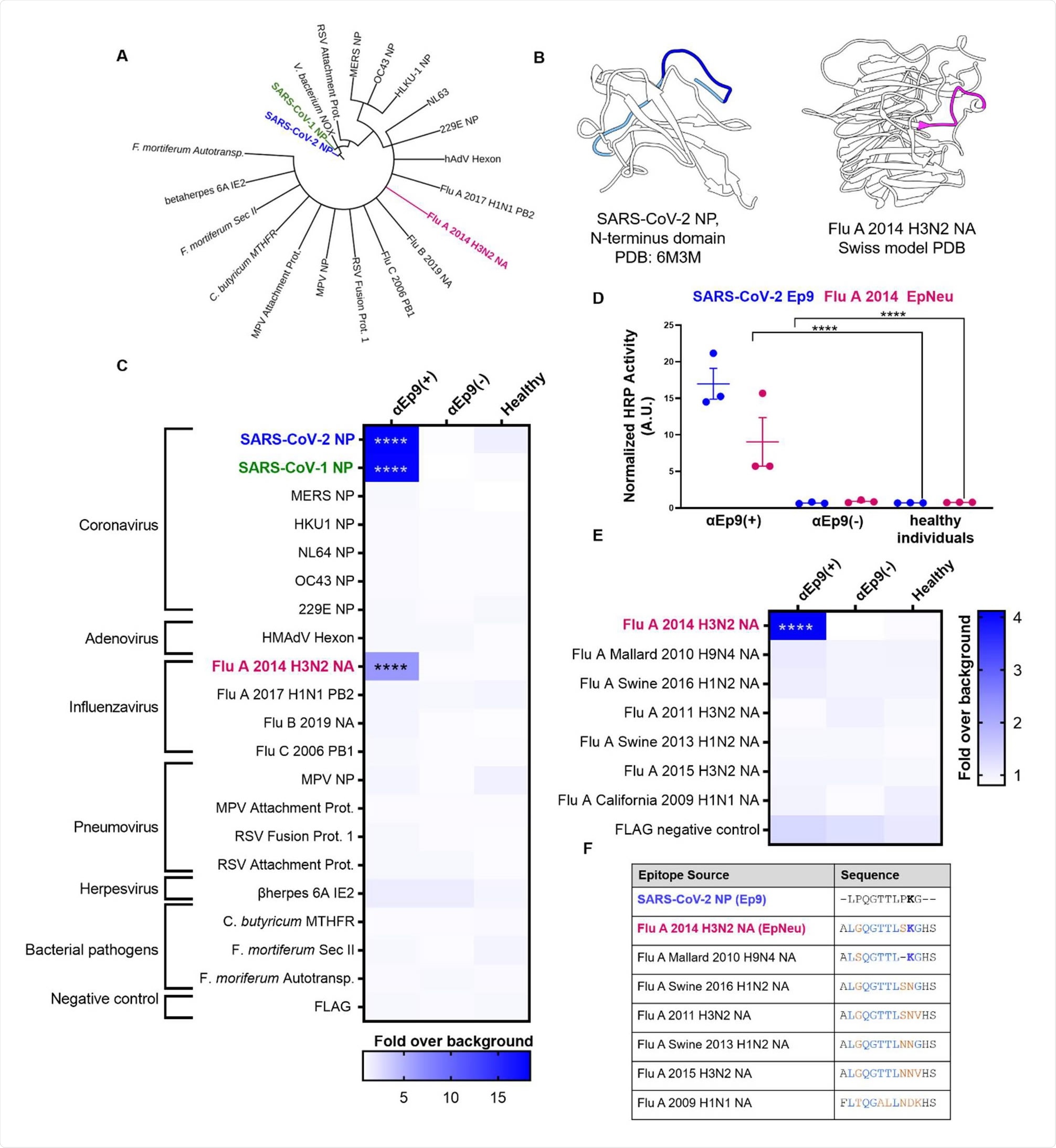
Potential OAS epitopes for binding αEp9 Abs suggested by bioinformatics and tested by phage ELISA. (A) Cladogram depicting sequence homology of the Ep9 sequence from SARS-CoV-2 to the bioinformatics-identified, closest homologs. Sequence alignments used pBLAST and VAST, and the cladogram was generated by iTOL (Letunic & Bork, 2019). (B) Structures of SARS-CoV-2 NP RNA binding domain (PDB: 6M3M) and the Flu A 2014 H3N2 NA protein (modeled by SWISS-Model (Waterhouse et al., 2018)). SARS-CoV-2 NP highlights Ep9 residues (light and dark blue) and the region homologous region to EpNeu (dark blue). The depicted model of Flu A 2014 H3N2 NA highlights the EpNeu putative antigen (pink). (C) ELISAs examined binding of phage-displayed potential OAS epitopes to Abs from three sets of pooled plasma from five αEp9(+) patients, or five αEp9(-) patients. Pooled plasma from healthy patients was an additional negative control. The colors of the heat map represent the mean binding signal normalized to phage background negative controls (signal from phage without a displayed peptide). (D) Expansion of data from panel C shows ELISA signals from the independently assayed individual pools shows results from the individual pools (****p <0.0001 for a two-way ANOVA comparing binding of phage-displayed epitopes listed in panel C to different groups of pooled plasma, ad hoc Tukey test). (E) Using EpNeu as the search template to generate homologous sequences (shown in next panel), ELISAs examined EpNeu homologs’ binding to pooled plasma from αEp9(+), αEp9(-), or healthy individuals. The data are represented as described in panel C (****p <0.0001 for two-way ANOVA c phage-displayed epitopes, ad hoc Tukey and Dunnett’s test as shown). (F) Amino acid sequence alignment of the closely related Flu A NA homologs of EpNeu from pBLAST(Altschul et al., 1997). Blue and orange residues represent conserved and mismatched amino acids, respectively, relative to Ep9. Bolded residues are important for epitope recognition by αEp9 Abs. Here, the term Flu refers to influenza.
Binding assays showed cross-reactivity between antibodies binding to Ep9 and a sequence derived from the neuraminidase protein of H3N2 Influenza A virus.
“The results support the hypothesis that the αEp9 Abs found in severe COVID-19 disease may have originated from a primary infection with H3N2 influenza A,” writes the team.
The αEp9 antibodies likely following widespread influenza in 2014
Since the neuraminidase protein is not present in the influenza vaccine, the researchers say that the αEp9 antibodies were likely generated following widespread influenza infection in 2014.
Patient histories rarely include previous influenza infections, so this information was not available for the participants in this study.
The H3N2 2014 neuraminidase sequence was isolated from Para in Brazil and the strain’s spread in North America remains unknown.
“However, a severe outbreak of influenza A H3N2 was recorded; in 2014, an antigenic shift from the vaccine strain A/Texas/50/2012(H3N2) resulted in low vaccine efficiency (14%), and high levels of other strains spread,” writes Weiss and colleagues.
What are the implications for future research?
The researchers say this study offers a molecular mechanism for OAS underlying the high rate of severe COVID-19 in αEp9(+) patients.
“Future studies could examine the correlation between a country’s rate of the H3N2 2014 influenza and severe COVID-19,” they write.
Furthermore, health systems may have recorded primary infections in certain patients, and these records could also be examined for correlations, suggests the team.
“Examining epitope conservation and antibody cross-reactivity could predict OAS-based immune responses and disease outcomes in future infections,” writes Weiss and colleagues
“Identifying detrimental, benign or beneficial OAS pathways could also guide vaccine design for increased efficacy and reduced risk,” they conclude.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.