The appearance of SARS-CoV-2 viral variants in the ongoing pandemic, as well as the rise of spontaneous viral resistance to conventional antiviral drugs, underscore an urgent need to develop new classes of effective antiviral agents.
We know that this virus can evade antiviral immunity by successfully interfering with the synthesis of host proteins, messenger RNA (mRNA) stability, as well as protein trafficking. More specifically, the SARS-CoV-2 nonstructural protein 1 (Nsp1) can utilize its C-terminal domain to block the ribosome function and halt host protein synthesis.
Moreover, previous studies on SARS coronaviruses have also demonstrated that the 5’ untranslated region (5’ UTR) of coronaviral mRNAs is involved in protecting the virus against Nsp1-mediated inhibition of mRNA translation.
Nonetheless, it is still unclear how exactly SARS-CoV-2 outmaneuvers Nsp1-mediated inhibition of viral protein synthesis and whether the mechanism in play is actually amenable to therapeutic targeting.
This was a research question a group of scientists from Harvard Medical School, Boston Children’s Hospital, Harbin Institute of Technology, Massachusetts Institute of Technology, and the Ohio State University aimed to tackle.
A multifaceted methodological approach
To investigate the function of SARS-CoV-2 Nsp1 in inhibiting mRNA translation, the researchers have co-transfected a mScarlet reporter construct (which is a basic, constitutively red fluorescent protein) with maltose-binding protein (MBP)-tagged Nsp1 or the MBP control in different cell lines.
For that purpose, state-of-the-art immunofluorescence and quantitative microscopy methods have been pursued, while transfection experiments for microscopy have been done in at least three replicates.
Finally, to suppress viral translation, they have attempted to disrupt the function of a critical short stemloop 1 (SL1) in SARS-CoV-2 5’ UTR function with the use of antisense oligonucleotides that have been designed to target various SL1 regions.
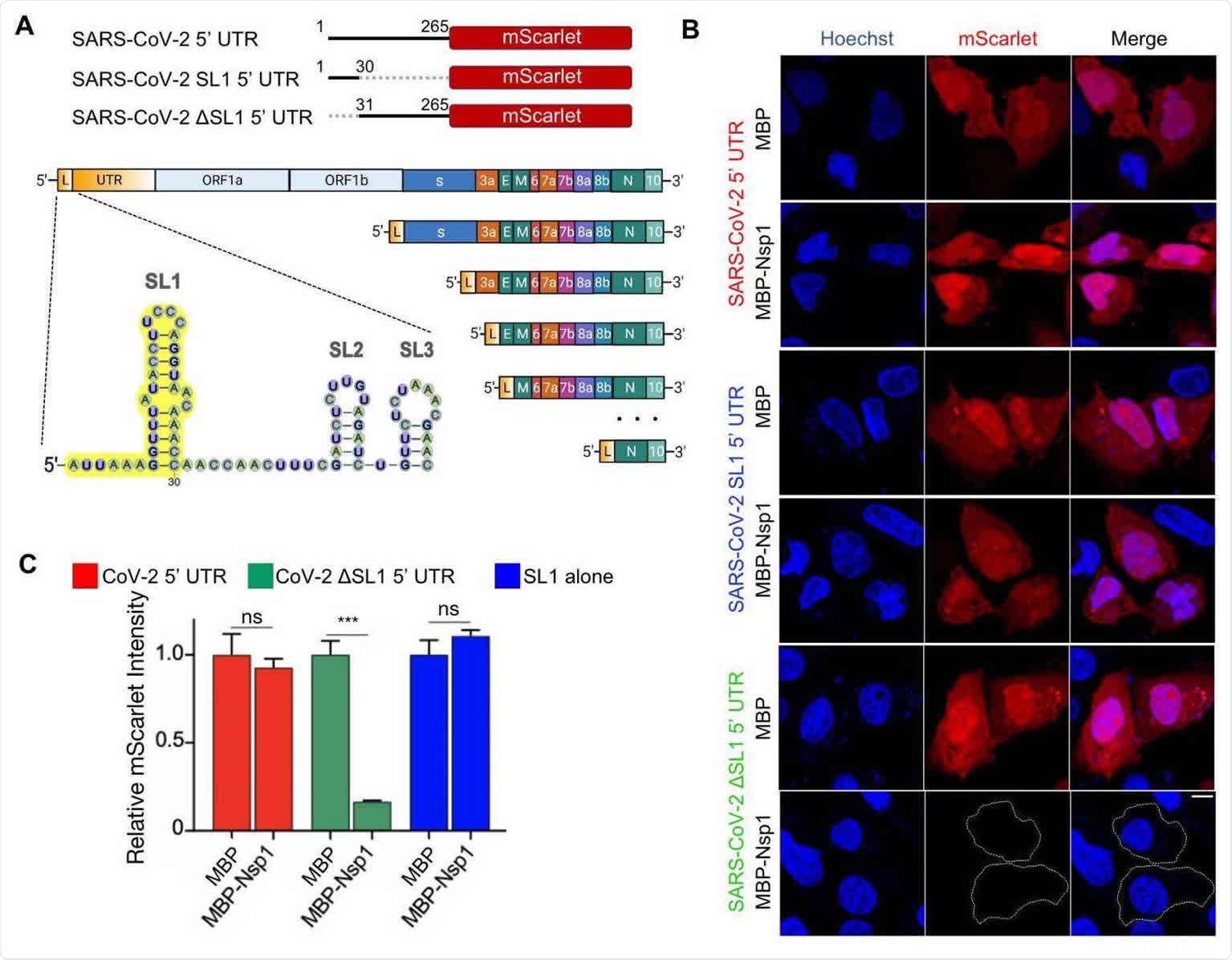
SARS-CoV-2 5’ UTR bypasses Nsp1-mediated inhibition of translation. (A) Schematic of translational reporters. 5’ UTR sequences from Control, MAVS, or SARS-CoV-2 were placed upstream of the mScarlet reporter (left panel). MBP or MBP-Nsp1 (right panel) were both downstream of Control 5’ UTR and were co-transfected along with each reporter plasmid. A CMV promoter was used to drive expression in all constructs. (B) Representative images of HeLa cells co-transfected with Control 5’ UTR reporter or SARS-CoV-2 5’ UTR reporter and either MBP alone or MBP-Nsp1 and visualized for DNA by Hoechst (blue), MBP by indirect immunofluorescence (green), and mScarlet by in situ fluorescence (red). Successfully transfected cells difficult to visualize due to low intensity are outlined here and in other figures. (C) Quantification of relative mScarlet intensity of data corresponding to (B). (D) Representative images of HeLa cells transfected with MAVs 5’ UTR reporter (red). (E) Quantification of relative mScarlet intensity in (D). (F) HeLa cells transfected with either ORF3a-GFP (left panel) or ORF8- GFP(right panel) downstream of SARS-CoV-2 5’ UTR. (G) Quantification of relative GFP intensity in (F). Error bars correspond to standard error of the mean except where otherwise noted. Scale bars are shown in each bottom right image and correspond to 10 microns.
Switching the translation machinery
This study has revealed that SARS-CoV-2 can utilize SL1 in its 5’ UTR to evade Nsp1 suppression and, in turn, effectively switch the translational machinery from synthesizing host proteins to making viral proteins. In that process, a certain level of host fitness is maintained while viral replication is maximized.
Moreover, the researchers have found that the N-terminal domain also contributes to suppressing host translation by coordinating with the C-terminal. Importantly, both domains have to be covalently linked and adequately spaced in order to perform this function in an optimal fashion.
Regarding therapeutic potential, it was shown that SL1 could be targeted by locked nucleic acid (LNA) antisense oligonucleotides in order to prevent SARS-CoV-2 5’ UTR from evading its own translational suppression to inhibit viral replication vigorously.
Implications for treatment
Since there are no known single nucleotide variants within the SL1 region with more than 1% in frequency, nor any known mutations among newly identified variants, this mechanism may actually represent a distinctive treatment target for increasingly infectious and immune-evasive strains that may continue to emerge during the pandemic.
“More generally, our proof-of-principle in developing therapeutics to unleash a pathogen’s own virulence mechanism upon itself may represent an important strategy to avoid resistance in SARS-CoV-2 and could be expanded to other host-pathogen systems”, say study researchers in this bioRxiv paper.
Future studies will be pivotal for teasing out specific functional interactions between Nsp1 and host proteins and whether mutants that could avoid such a therapeutic approach would be at a substantial selective disadvantage.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Vora, S.M. et al. (2021). Targeting Stem-loop 1 of the SARS-CoV-2 5’UTR to suppress viral translation and Nsp1 evasion, bioRxiv, https://doi.org/10.1101/2021.09.09.459641, https://www.biorxiv.org/content/10.1101/2021.09.09.459641v1
- Peer reviewed and published scientific report.
Vora, Setu M., Pietro Fontana, Tianyang Mao, Valerie Leger, Ying Zhang, Tian-Min Fu, Judy Lieberman, et al. 2022. “Targeting Stem-Loop 1 of the SARS-CoV-2 5′ UTR to Suppress Viral Translation and Nsp1 Evasion.” Proceedings of the National Academy of Sciences of the United States of America 119 (9): e2117198119. https://doi.org/10.1073/pnas.2117198119. https://www.pnas.org/doi/full/10.1073/pnas.2117198119.