Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection causes coronavirus disease 2019 (COVID-19), which has evolved into a global pandemic with over 240 million reported cases as of 18 October 2021.
The global pandemic has highlighted various health disparities that have impacted infection prevalence and vaccine uptake. For example, in the US, black and Latino individuals have been reported to have had higher rates of SARS-CoV-2 infection and mortality compared to white individuals.
Public health interventions to address these disparities are essential for any country to combat the pandemic effectively. Emergency departments (ED) provide good data on symptomatic COVID-19 infections. However, serological assessments may provide more accurate information on SARS-CoV-2 seroprevalence, which may help identify health disparities.
A new study recently published on the medRxiv* preprint server demonstrates the use of a serological testing algorithm for identifying disparities in infection rates and vaccine uptake amongst the study population to enable public health interventions.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Validation of the algorithm
Training and validation of algorithms are done using sample data, and then they are applied to clinical practice. Validation is required, both for sensitivity and specificity (ability to detect positives and negatives).
The algorithm generated in this study was validated using three sample sets comprising subjects with a previous infection history and known to have received SARS-CoV-2 vaccination. Samples with known vaccination history included subjects from a vaccine phase I trial and hospital healthcare professionals (HCP). Samples from individuals who have previous SARS-CoV-2 infection were derived from convalescent plasma donors (CCP), clinical and characterization protocol for severe infectious diseases (CCPSEI), and HCP who have positive SARS-CoV-2 RT-PCR test results. In addition, specificity was validated using pre-pandemic remnant complete blood count (CBC) samples collected from Johns Hopkins Hospital Emergency Department (JHH ED) patients.
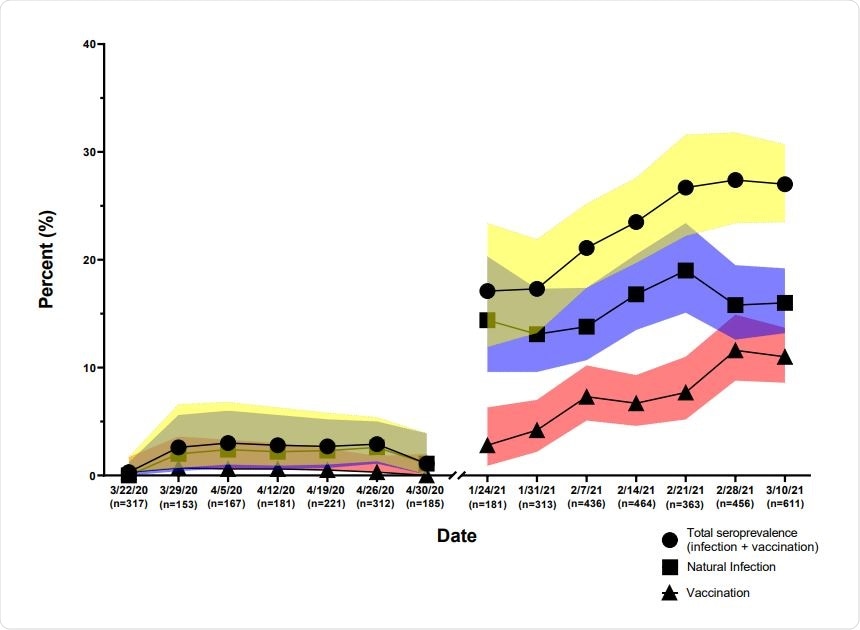
Seroprevalence of antibodies of SARS-CoV-2 2020-2021. JHHED ED samples from 2020 and 2021 were tested on the previously mentioned algorithm and categorized according to the date on which the sample was drawn.
Application of the validated algorithm
After validation, the algorithm was applied to two serosurveys conducted on 145 patients who attended the JHH ED in Baltimore city from 16 March to 30 April 2020 and from 11 January to 10 March 2021.
During the study period of the surveys, remnant CBC blood samples were collected from patients aged more than 17 years who visited the ED. Each sample was assigned a unique study code, and protected health information was de-identified.
Data on the COVID-19 vaccination status of the study population was not available. Therefore, the demographic dataset containing information on age, sex, ethnicity, etc., was de-linked from the samples, and laboratory tests for determining the SARS-CoV-2 serostatus were performed. After the tests were completed, the serostatus of each sample was then linked to the demographic data set using the assigned study code.
Serological assays to detect serological reactivity towards SARS-CoV-2 spike, spike glycoprotein receptor binding domain and nucleocapsid
Three serological assays were selected that will help differentiate serological reactivity towards SARS-CoV-2 spike (S1), spike glycoprotein receptor-binding domain (RBD), or nucleocapsid:
Euroimmun Anti-SARS-CoV-2 ELISA quantifies the IgG responses to SARS-CoV-2 S1. Bio-Rad Platelia SARS-CoV-2 Total Antibody ELISA measures total antibodies elicited against the SARS-CoV-2 nucleocapsid. In the case of both the ELISA serology tests, the results are generated as optical density of the sample divided by the control (S/C). If S/C ≥ 0.8 the sample was considered as positive. CoronaCHEKTM COVID-19 IgG/IgM Rapid Test Cassette detects the presence of IgM and IgG against the RBD of the spike protein. The presence of a visible band is considered as positive.
In the study, an algorithm made of the Euroimmun, Bio-Rad, and CoronaCHEK assays was employed to differentiate samples and divide them into:
The naturally infected group comprising of subjects who may or may not be vaccinated with prior infection history. Samples that showed a positive or indeterminate result on the Bio-Rad assay were segregated to this group.
The vaccinated group consisting of subjects who are vaccinated and who do not have any prior infection history. Samples that were positive on CoronaCHEK and negative on Bio-Rad were separated in this group.
The unexposed group, neither vaccinated nor prior infections. False-positive samples tested positive in the Euroimmun assay but negative on CoronaCHEK were segregated to this group.
The algorithm was validated by assessing its diagnostic accuracy by using samples of known status, e.g., to evaluate if the algorithm can detect vaccinated samples. It was tested on samples known to belong to the vaccinated group.
The algorithm exhibited good specificity and sensitivity in detecting and differentiating the samples
Upon validating the algorithm, it was found that it showed 100% sensitivity and specificity for detecting vaccinated samples. Furthermore, in the case of naturally infected samples, it was able to differentiate them with 84.4% and 100 % sensitivity and specificity, respectively.
Amongst the cohorts used for algorithm validation, considerable differences were detected between their serostatus and their antibody reactivity levels to spike and nucleocapsid.
The median S/C value for reactivity towards spike was 8.9 and 5.2 in the case of vaccinated and infected individuals, respectively showing a clear difference in levels of reactivity.
Interestingly, in the case of median S/C for antibody reactivity to nucleocapsid, it was found that in vaccinated persons with no prior infections, the value did not cross the threshold value of 0.8 for a positive result. At the same time, the naturally infected group who were never vaccinated showed an S/C of 4.3.
Among the HCP, the individuals who had a known PCR positive test and were subsequently vaccinated showed a spike antibody median S/C value similar to the vaccinated group (median=9.5) and nucleocapsid antibody median S/C value that was similar to the naturally infected group of individuals (median=3.4).
In the case of HCP who were SARS-CoV-2, PCR negative with suspected infections showed median S/C values for spike antibody (median=10.0) and nucleocapsid (median=3.3) similar to individuals with known infection who were subsequently vaccinated.
Very little reactivity was found in the case of the pre-pandemic samples.
An increase in seroprevalence and vaccinated individuals was observed during the study period of 2020 and 2021
The testing algorithms were applied to two serosurveys JHH ED 2020 and JHH ED 2021. During the combined study period of the two surveys, seroprevalence increased from 1.6% to 23.8%. It was found that during the second survey, the prevalence of vaccinated individuals increased from 2.8% to 11% during the period between mid-January to mid-March of 2021.
Demographic characteristics like age, sex, race, and ethnicity were similar for the two survey periods. The Infection prevalence during the Spring of 2020 was not found to vary with age, sex, race or ethnicity. However, significant variation was found in the infection prevalence based on these demographic characteristics during the Spring of 2021.
Disparities detected in SARS-CoV-2 prevalence and vaccine uptake
The presence of antibodies to SARS-CoV-2, which is indicative of prior infections or vaccinations, was assessed. In 2020 and 2021, it was found from the surveys that white men and women had the lowest prevalence of infections.
Among white women, the proportion of vaccinated individuals was higher than infected individuals, which was not observed in any other group. In addition, amongst the study subjects, by Spring 2021, Hispanics were found to have the highest percentage of previous SARS- CoV-2 infection compared to other ethnic groups.
No statistical difference in infection rates based on age was found in the 2021 survey. However, it was found that 45-59-year-olds were less likely to be vaccinated compared to the younger age group. In addition, white women are less likely to be previously infected compared to Black women and Hispanic men and women. It was also observed that White men and women and Hispanic men were more likely to be vaccinated when compared to black women.
The data from the surveys were further stratified based on sex, race, and ethnicity. It was found that individuals between the ages of 45 – 74 were less likely to have developed natural infections when compared to the younger age group of 18 – 29.
White individuals were less likely to be infected compared to the Black population. Women had an increased odds of being vaccinated when compared to men after adjusting for age, race and ethnicity.
Further, subjects in the age group of 45 -59 had a lesser likelihood of being vaccinated compared to the 18 – 29 age group. Further, ethnicity did not significantly influence differences in vaccinations.
Conclusion
The study did have several limitations: individuals with known breakthrough infections were not tested, naturally infected individuals who were not vaccinated were not differentiated from those who were vaccinated, and lack of seroreactivity was seen in a small number of naturally infected individuals. However, the algorithm employed in this study efficiently differentiated naturally infected, vaccinated, and unvaccinated individuals.
The present study highlights health disparities in the Baltimore city population, showing that differences based on gender, race, and ethnicity exist in SARS-CoV-2 infection prevalence and vaccine uptake.
The findings from this study demonstrate that in cases where the history of vaccination and infections is not available serologic testing algorithms could be used to differentiate the populations in order to facilitate public health interventions.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Beck, E. J. et al. Differentiation of SARS-CoV-2 naturally infected and vaccinated individuals in an inner-city emergency department, doi:10.1101/2021.10.13.21264968, https://www.medrxiv.org/content/10.1101/2021.10.13.21264968v1
- Peer reviewed and published scientific report.
Beck, Evan J., Yu-Hsiang Hsieh, Reinaldo E. Fernandez, Gaby Dashler, Emily R. Egbert, Shawn A. Truelove, Caroline Garliss, et al. n.d. “Differentiation of Individuals Previously Infected with and Vaccinated for SARS-CoV-2 in an Inner-City Emergency Department.” Journal of Clinical Microbiology 60 (3): e02390-21. Accessed November 24, 2022. https://doi.org/10.1128/jcm.02390-21. https://journals.asm.org/doi/10.1128/jcm.02390-21.