The coronavirus disease 2019 (COVID-19) is caused by the SARS-CoV-2 infection. As of November 11, 2021, over 250 million cases of SARS-CoV-2 infection have been reported worldwide, which has resulted in over 5 million deaths.
Despite mitigation strategies and vaccination rollouts, SARS-CoV-2 continues to spread and cause reinfection in vaccinated individuals as a result of emerging variants across countries. Therefore, it is imperative to develop an accessible, early, and accurate diagnostic method to facilitate robust public health surveillance and rapid testing.
Shortcomings of the current gold-standard technique for the SARS-CoV-2 testing, the reverse transcription-polymerase chain reaction (RT-PCR), include requirements for highly trained personnel, dedicated facilities, and instrumentations.
Comparatively, the RT-LAMP method is an isothermal amplification technique that has been widely studied as an alternative for COVID-19 testing. This technique serves as a potential point-of-care tool because it is a rapid, sensitive, and specific technique.
Extensive designing, validation, and optimization of the technique have been completed, coupled with diverse optical detection readout methods such as changes in turbidity caused by magnesium pyrophosphate precipitate, changes in fluorescence using dyes, colorimetric pH indicators, or gel electrophoresis followed by UV detection.
While each of these methods would require a minimum concentration of the signaling reporters (cmin) for the readout system to distinguish between SARS-CoV-2 positive and negative samples, the concentration of the cmin could range from millimolar (mM) to nanomolar (nM).
Depending on the average reaction rate of the assay, the required minimal concentration can be related to the time of the reaction. Thus, reducing the detection cmin is preferred towards quick time to positive.
In an effort to achieve this, the intrinsic single-molecule sensitivity of nanopore sensors is useful, as these sensors significantly reduce the cmin. In fact, this combined approach can allow the the analyte concentration to be within the picomolar (pM) range, thus reducing the time required to determine whether the test is positive or negative.
In a previous study, a LAMP-coupled nanopore sensor for malaria nucleic acid test was demonstrated. Based on label-free electronics-based nanopore sensors, this method is a new avenue for making accessible molecular testing at the point of care. In the present study, the researchers report using the RT-LAMP coupled nanopore platform for rapid detection of the SARS-CoV-2.
About the study
Using spiked saliva samples and COVID-19-positive clinical nasopharyngeal swab samples, the researchers established detection of SARS-CoV-2 using the RT-LAMP coupled nanopore platform.
Here, the nanopore sensor detects the amplicon size and measures the concentration by the digital counting method for qualitative assessment of positive/negative results. The researchers then compared the time to determine whether the sample was positive and assess the sensitivity performances of two one-pot RT-LAMP assays targeting the nucleocapsid (N) and envelope (E) genes of SARS-CoV-2.
The researchers presented the workflow of the RT-LAMP coupled nanopore method for SARS-CoV-2 detection from sample collection, preparation, to RNA extraction from the samples. The RT-LAMP amplification reaction is performed at 65◦C for 15 min. During the nanopore readout in a negative control sample, amplification is negligible, whereas in a positive case, amplicons are increased significantly, resulting in a sharp increase in event rate.
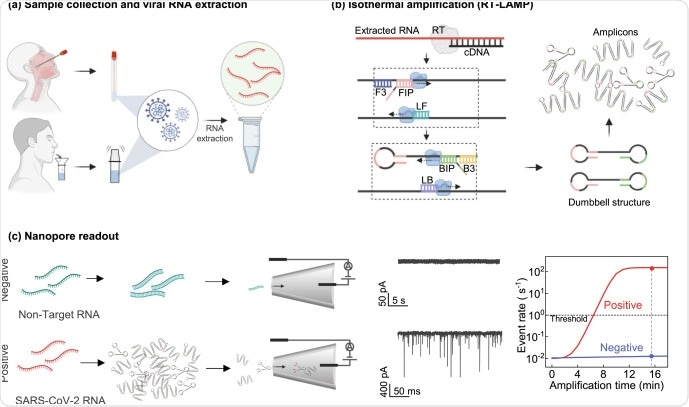 Workflow of RT-LAMP coupled nanopore method for SARS-CoV-2 detection. (a) Sample collection, preparation, and RNA extraction from either the nasopharyngeal swab sample or the saliva sample. (b) RT-LAMP amplification. One step RT-LAMP reaction is performed at 65°C for 15 min. (c) Nanopore readout. In a negative control sample, no amplification occurs, resulting in a negligible event rate. In a positive case, ampliconsincreased significantly, resulting in a sharp increase in event rate. The right panel shows the nanopore event rate as a function of RT-LAMP reaction time. The event rate threshold was set at 1s-1 as the criteria for a positive call.
Workflow of RT-LAMP coupled nanopore method for SARS-CoV-2 detection. (a) Sample collection, preparation, and RNA extraction from either the nasopharyngeal swab sample or the saliva sample. (b) RT-LAMP amplification. One step RT-LAMP reaction is performed at 65°C for 15 min. (c) Nanopore readout. In a negative control sample, no amplification occurs, resulting in a negligible event rate. In a positive case, ampliconsincreased significantly, resulting in a sharp increase in event rate. The right panel shows the nanopore event rate as a function of RT-LAMP reaction time. The event rate threshold was set at 1s-1 as the criteria for a positive call.
The nanopore event rate has a linear relationship with the analyte concentration. The researchers reported that the event rate threshold was set at 1s-1 as the criteria for a positive result. Notably, this threshold is much higher than the background event rate in the negative control (<0.029 s-1), such that the false-positive rate can be minimized.
Further, the researchers validated the method by adopting two LAMP primer sets targeting the N and E genes of SARS-CoV-2, respectively. They demonstrated that the N assay was quick to reach a positive result and had better copy number sensitivity.
Therefore, they chose the N primer set for our SARSCoV-2 RT-LAMP assay in this study. The researchers optimized the RT-LAMP assay targeting the N gene to show a limit of detection (LoD) of 65 copies at the 95% confidence level.
Significantly, the test is also highly specific against other human coronavirus targets.
The researchers performed a nanopore counting analysis on the resulting amplicons, examined by the glass nanopore sensor, and benchmarked the event rate with a threshold. The method was quicker because of its intrinsic single-molecule level of sensitivity.
Further, using real samples, the researchers demonstrated that the results confirmed an excellent analytical specificity of the RT-LAMP coupled nanopore sensor against the SARS-CoV-2.
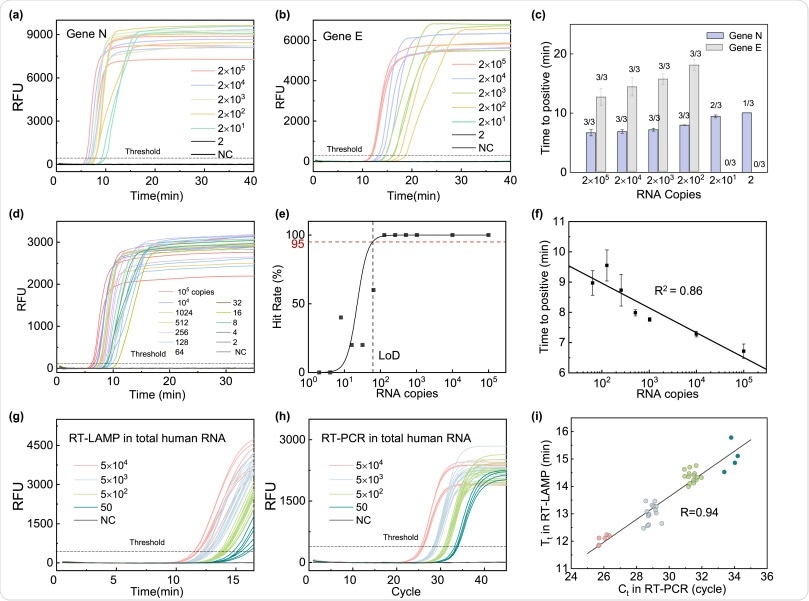 RT-LAMP assay validation. (a) N primer set results, and (b) E primer set results with viral RNA concentrations ranging from 2 and 105 copies per reaction. (c) Time to positive value comparison between the N primer set (blue bars) and the E primer set (grey bars) at different RNA concentrations. The N primer set showed better performances in terms of sensitivity and time to positive. (d) Real-time RT-LAMP result with a finer serial dilution (2 × ) using N primer set. (e) The extracted hit rate at various RNA concentrations to establish the assay LoD, which is determined to be 65 copies at 95% confidence level. (f) Time to positive value with N primer sets at concentrations ranging between 102 and 105 copies per reaction. A good linearity is obtained, indicating that a semi-quantitative test is feasible. (g) Real-time RT-LAMP result in saliva RNA background. (h) Real-time RT-PCR result in saliva RNA background. (i) The correlation between the RT-PCR and RT-LAMP measurement in total saliva RNA background. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
RT-LAMP assay validation. (a) N primer set results, and (b) E primer set results with viral RNA concentrations ranging from 2 and 105 copies per reaction. (c) Time to positive value comparison between the N primer set (blue bars) and the E primer set (grey bars) at different RNA concentrations. The N primer set showed better performances in terms of sensitivity and time to positive. (d) Real-time RT-LAMP result with a finer serial dilution (2 × ) using N primer set. (e) The extracted hit rate at various RNA concentrations to establish the assay LoD, which is determined to be 65 copies at 95% confidence level. (f) Time to positive value with N primer sets at concentrations ranging between 102 and 105 copies per reaction. A good linearity is obtained, indicating that a semi-quantitative test is feasible. (g) Real-time RT-LAMP result in saliva RNA background. (h) Real-time RT-PCR result in saliva RNA background. (i) The correlation between the RT-PCR and RT-LAMP measurement in total saliva RNA background. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Conclusions
The current study reports an RT-LAMP coupled nanopore platform for rapid detection of SARS-CoV-2. Compared to bulk conventional optical techniques, this nanopore sensor enables one to identify positive/negative samples faster, with 98% diagnostic sensitivity and 92% diagnostic specificity compared to RT-PCR.
Employing this RT-LAMP coupled electronic nanopore digital counting platform technique for SARS-CoV-2 detection has significant potential to make next-generation accessible diagnostic testing that is rapid, sensitive, and specific.