Even as scientists around the world have made efforts to develop and produce safe and effective vaccines to counter the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the agent that caused the ongoing pandemic of coronavirus disease 2019 (COVID-19), this virus continues to mutate. Soon after the rollout of the first vaccines from Pfizer and Moderna, both of which are built on the messenger ribonucleic acid (mRNA) platform, the emergence of the Delta variant took the world by storm, driving up cases, deaths, and hospitalizations to horrifying levels worldwide.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The most recently identified SARS-CoV-2 variant is the Omicron variant (B.1.1.529). This variant, which was rapidly named a variant of concern (VOC) by the World Health Organization (WHO), has the highest number of mutations of all known variants; however, their biological impact is still to be revealed.
A new study published on the bioRxiv* preprint server discusses the effect of the Omicron variant’s mutations on its binding efficiency to the angiotensin-converting enzyme 2 (ACE2) receptor, which mediates viral attachment and entry into host cells.
Background
The rapid rate at which infections due to the Omicron VOC of SARS-CoV-2 are rising after its recent emergence has caused alarm to public health authorities and governments around the world. Almost by reflex, many countries immediately blacklisted flights and other routes of entry from numerous countries in a futile attempt to reduce the transmission of this new variant.
The Omicron VOC has 30 mutations on the spike glycoprotein, of which 14 are on the receptor-binding domain (RBD) of the spike protein. The RBD is crucial to virus-receptor binding via its interactions with the peptidase domain (PD) of the ACE2 receptor.
The multiple new and shared mutations on the spike glycoprotein, especially the RBD, of the Omicron variant causes differences in the way it interacts with the ACE2 PD and thus affects the rate and ease of viral entry. The RBD of the spike protein is the target of neutralizing antibodies elicited by natural infection or vaccination with all currently available vaccines.
The mutations seen on the RBD of this new variant are exposed on the surface and are targeted by antibodies and nanobodies such as H11-H4, H11-D4, and Ty1 that are currently being developed as potential therapeutic agents against COVID-19. Of the 15 RBD mutations, 11 are at the ACE2-PD interface. Five of these mutations were previously shown by molecular dynamics (MD) simulations to interact with corresponding residues on the PD in the wild-type virus.
These interactions include salt bridges formed between the K417-D30 and E484-K31, as well as hydrogen bonding between Q493-E35, Q498-Q42, Q498-K353, and Y505-E37. At present, there is not enough data to specify the effect of the Omicron mutations on the binding strength between the Omicron RBD and ACE2. It is also currently unclear whether antibodies elicited by earlier variants of SARS-CoV-2 will be able to neutralize the ACE2-RBD interactions and thus prevent virus entry.
In order to help answer this question, the researchers carried out all-atom simulations of the PD under appropriate conditions.
Study findings
The simulations were carried out in the presence of a total of 200,000 explicit water and ions, as well as the full-length sugar molecules on both the RBD and ACE2 molecules. A total of 900 nanoseconds (ns) of simulation length was obtained using the same methods used earlier to derive MD simulations of the wildtype strain and both the Alpha, and Beta VOCs.
The researchers found that the interactions between the Omicron RBD and the ACE2 PD were much more extensive compared to the wild-type RBD. This is due to an increase in the total number of salt bridges by 250%, with seven new salt bridges being formed with the loss of two from the wildtype RBD-ACE2 interaction network.
Meanwhile, the ten hydrophobic interactions formed by the wildtype variant were conserved, while one more was added between Y501-Y41 for an increase of 10%. The hydrogen bonding at the receptor-spike interface was decreased from the wildtype, which showed eight bonds, six of which were with the Omicron variant, for a total reduction of 10%. Interestingly, all but one of these six bonds were newly formed hydrogen bonds that were not observed in the wildtype RBD-ACE2 interactions.
The conformation-based binding energies at the receptor-binding interface for the two variants were estimated by the Molecular Mechanics Poisson-Boltzmann Surface Area (MMPBSA) method. When coupled with the four sets of simulations of the RBD-PD of the Omicron variant, this method showed an increase in binding energy by 44% for the Omicron variant as compared to the wild-type strain of SARS-CoV-2.
With the wildtype variant, the salt bridges occurred mostly at the interface of the contact regions (CR) CR1 and CR2, with most of the hydrogen bonds at CR3 and hydrophobic bonds at CR1. The Omicron RBD mutations have led to a more extensive and widely distributed network of bonds that extend to both sides of the interaction surface.
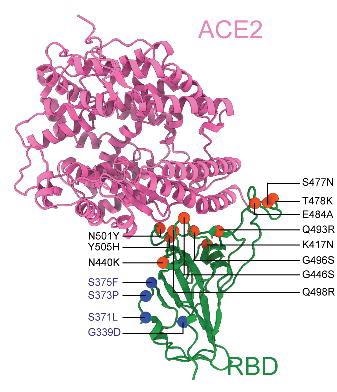 Location of RBD mutations for the Omicron variant.
Location of RBD mutations for the Omicron variant.
Implications
The results of the simulations and calculations presented in the current study demonstrate that the Omicron RBD may bind the ACE2 receptor with greater efficiency and thus infect the host cells more readily. More specifically, the network of lateral interactions around the interface of the RBD-PD is spread out over a larger area for the Omicron variant as compared to the wild-type strain.
Secondly, the mutations in the Omicron variant appear to lead to an altered distribution of the network of interactions between the virus RBD and its binding receptor, the ACE2 molecule, at the PD.
The changes in the area of interaction and the types and positions of bonds could potentially change the way the virus binds to the receptor. It could also affect neutralization by therapeutic or vaccine-induced neutralizing antibodies and nanobodies.
For instance, the E484K mutation could disrupt the formation of the salt bridge between E484-R52 and the hydrogen bond between E484-S57 by therapeutic agents such as H11-H4 or H11-D4. The same mutation may also eliminate the E484-N56 and E484-Y335 hydrogen bonds in Ty1.
This would prevent efficient binding and neutralization of the virus by these antibodies and nanobodies. In addition, the presence of the Q493R mutation could lead to the absence of the hydrogen bonds Q493-Y104 and Q493-S104 in H11-H4, and H11-D4, respectively.
It is clear that the impact of these and other mutations on the susceptibility of the Omicron VOC to antibody-mediated neutralization must be clarified in future research.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources