SARS-CoV-2 has a large genomic constitution encompassing single-stranded ribonucleic acid (RNA) and 12 open reading frames (ORFs) encoding for 26 proteins of structural, non-structural, and accessory types. The increased plasticity and resiliency of accessory proteins enhance the susceptibility of the accessory genes to undergo conformational changes such as insertions and deletions.
The deletions have given rise to mutant accessory variants that enhance viral ability to escape the protective immune mechanisms of the host, thereby increasing viral transmission. Nucleotide alterations lead to amino acid (aa) replacements in the viral molecule. In addition to these configurational substitutions, the insertions and deletions generate mutant variants that lead to the emergence of new viral strains with improved viral infectivity and diversified clinical manifestations.
The ORF7a gene belongs to the N.7 lineage, derived from the B.1.1.33 lineage prevalent in the early COVID-19 phase in Brazil. ORFs encode transmembrane proteins localized in the endoplasmic reticulum and Golgi network. After fusion of ORF7a and ORF7b variants into the OFR7ab strain, the viral cells attach to monocytes and reduce their antigen-binding ability along with an overproduction of inflammatory mediators such as cytokines.
Previous studies have established that recurring deletions in the terminal region of the spike (S) viral protein and ORFs of accessory genes contribute to immune evasion and confer resistance to the virus against host antibodies. However, the data is underestimated. Hence, the authors of the current study assessed how genetic variability led to deletions in the ORF7ab mutant variant affects the structure, pathogenicity, and clinical severity of COVID-19.
About the study
In the present study, 51 patients infected with COVID-19 between July 2020 to October 2020 were screened. Thirteen samples comprising oropharyngeal and nasopharyngeal swabs were collected from these patients, from which RNA was extracted to synthesize complementary DNA (cDNA) via real-time reverse transcription-polymerase chain reaction (RT-qPCR).
A genomic library was prepared, after which about 500 genomic variants were sequenced and mapped. In addition, the newly sequenced genomes of N.7 lineage were added to the Uruguayan genomes. cDNA was analyzed for the presence of genetic alterations such as single (12 nucleotides variant or ORF7a) deletion and double (12 nucleotides and 68 nucleotides fused variant or ORF7ab) deletions.
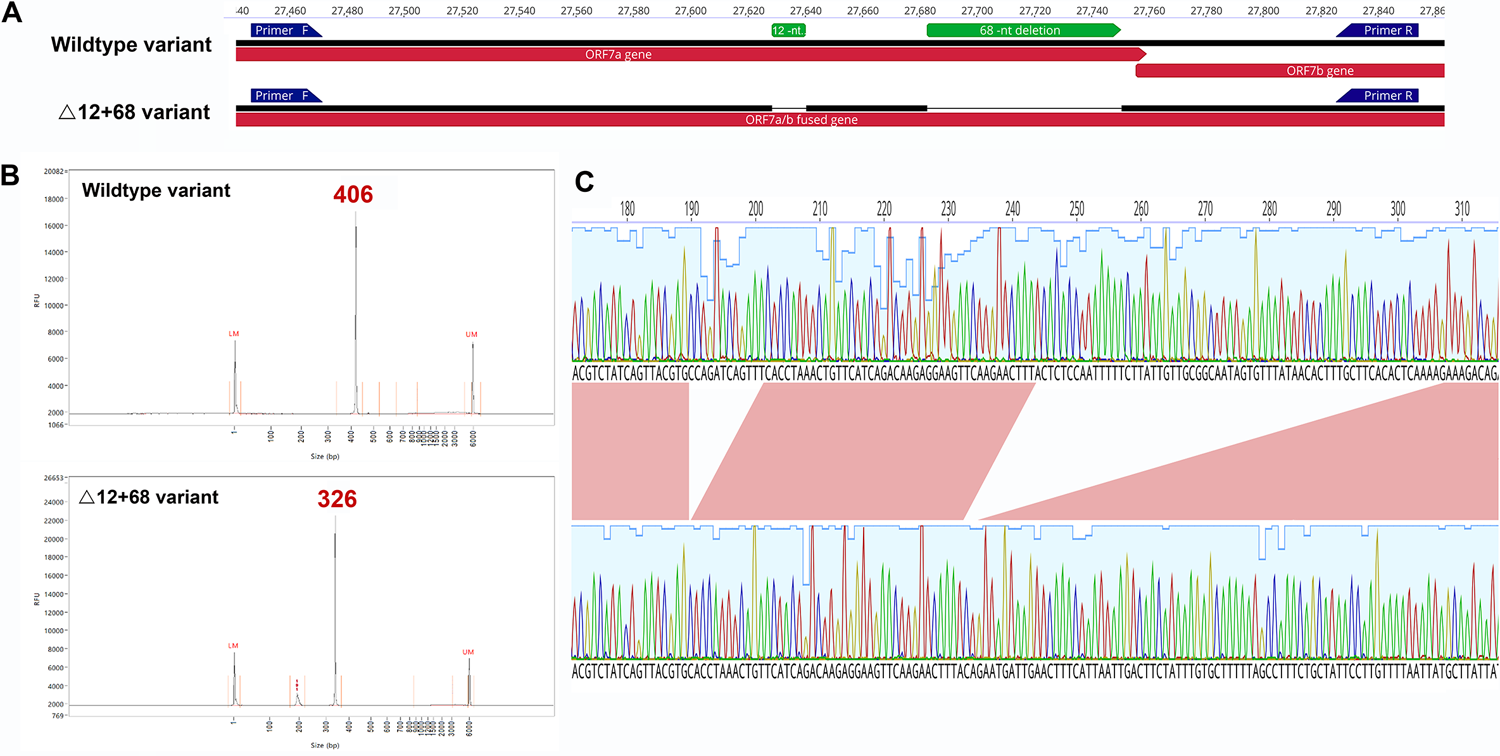
A: ORF7a amplicon for the wildtype and Δ12+68 variants. Artic primers annotations are visualized by blue arrows. B: Chromatogram peaks after capillary electrophoresis, wildtype, and Δ12+68 variants. C: Zoomed diagram details nucleotide sequence of the double deletion obtained by Sanger sequencing. https://doi.org/10.1371/journal.pone.0263563.g001
Results and discussion
Out of the 13 samples, nine genomic samples showed ORF7a deletions, whereas four samples showed both deletions. The mutant ORF7ab variant contained alterations in the stop codon sequence. The novel frame-shifting 68 nucleotide deletion was located 44 nucleotides away within the terminating domain of the ORF7A variant. All N.7 lineage mutations with deletions showed aa substitutions in R36Q and L37F genomes. The ORF7a variant has four aa deletions in positions 80 to 83 that fuses with the ORF7B variant with deletions in 23 aa at post-96 positions in the 138-codon sequence.
The researchers also evaluated the relationship between viral genomic deletions and the clinical presentation of COVID-19. Patients with deletions exhibited several clinical signs and symptoms along with two deaths, whereas patients with double deletions were either asymptomatic or had mild infections but fully recovered from COVID-19 post-treatment.
Conclusion
In the present study, the generation of the fused ORF7ab variant due to consecutive gene deletions gave rise to a mutant protein that could mimic normal transmembrane proteins and thus escape host immune mechanisms. The emergence of such a mutant could significantly enhance the transmission of COVID-19. The clinical correlation between genetic variability and gene deletions indicated that single-gene deletion was deleterious, whereas double gene mutation had a protective action concerning disease severity and recovery.
According to the team, this was the first case of consecutive gene deletions in SARS-CoV-2. However, future studies with larger sample sizes, genomic analysis of novel strains such as Zeta (P.2) strains and Gamma (P.1) derived from the same B.1.1.28 lineage and fitness analysis to determine the individual effects of the novel 68 nucleotide variant on the clinical course of COVID-19 must be carried out to devise genomic strategies to combat COVID-19.