Scientists have worked extensively hard to understand all aspects of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causal agent of the ongoing coronavirus disease 2019 (COVID-19) pandemic. SARS-CoV-2 is an RNA virus belonging to the family Coronaviridae of the genus Betacoronavirus. This virus invades the host cell via its surface spike (S) protein, which binds to the angiotensin-converting enzyme 2 (ACE2) of the host.
Previous studies have shown that proteolysis does not occur for proteins present on the virus's surface or the host cell surface. However, membrane-bound proteases, for example, 'a disintegrin and metalloproteases' (ADAMs), TMPRSS2, and beta-site APP cleaving enzymes (BACEs) can eliminate the ectodomain of surface proteins.
Several studies have shown that shed particles contain virus receptors, for example, Coxsackievirus and Adenovirus Receptor, the SARS-CoV-2 coreceptor NRP1 receptor, ACE2, the enterovirus 71 receptor P-selectin glycoprotein ligand 1 (PSGL-1), etc. The presence of other host cell shedding proteases that contribute to infection may be targeted in order to inhibit the spread of SARS-CoV-2.
Although several studies have indicated that exogenous addition of proteases (e.g., trypsin or overexpressed TMRPSS2) can increase the formation of syncytia in vitro, it is not evident if endogenous shedding proteases are associated with a similar mechanism. Therefore, it is unclear whether endogenous shedding proteases can be used as a potential pharmaceutical target to inhibit pathological syncytia formation.
A New Study
A new study published in the journal EMBO Reports has reported on the shedding of two proteases, namely, ADAM10 and ADAM17, which aids in S protein-dependent molecular events. Research has shown that ADAM10 and ADAM17 are essential for SARS-CoV-2 pathogenesis in terms of COVID-19 infection and SARS-CoV-2-mediated lung cell fusion.
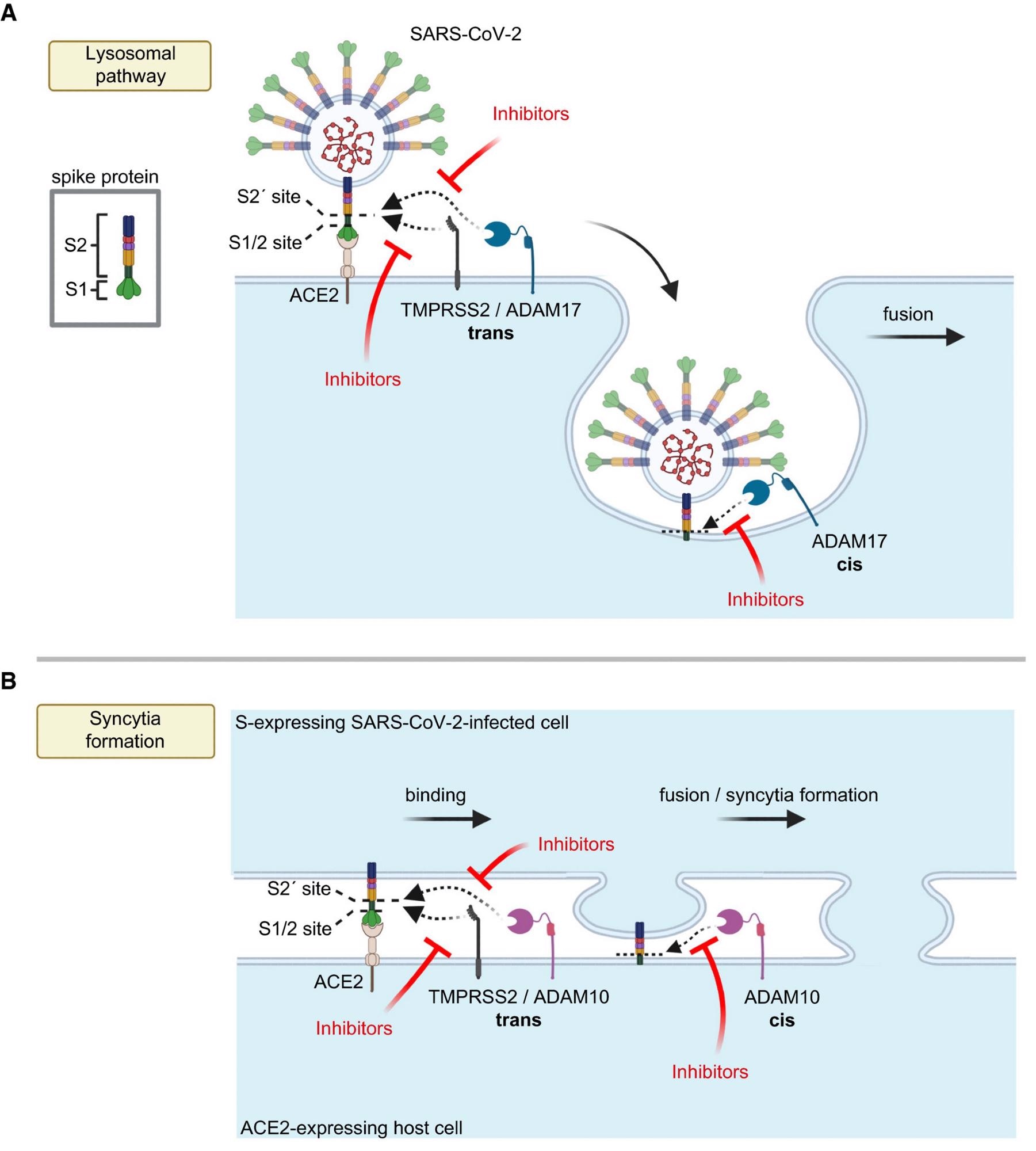 Model for the role of ADAM proteases in SARS-CoV-2 viral entry and lung cell syncytia formation A, B. The SARS-CoV-2 spike protein S consists of the two subunits S1 and S2 and binds through S1 to ACE2 on host cells, which is required for viral fusion with host cells (A) and for syncytia formation (B). (A) TMPRSS2 may cleave S in trans and allow for the fusion of SARS-CoV-2 with the host cell plasma membrane. In contrast, ADAM17 may either cleave S in trans at the plasma membrane or may cleave it once the fusion peptide of S has inserted into the host plasma membrane after endocytosis, which is conceptually similar to a cleavage in cis. After ADAM17 cleavage, the fusion of SARS-CoV-2 with the host cell is likely to happen after endocytosis. Inhibition of ADAM17 reduces viral uptake and fusion. (B) For syncytia formation, the S protein expressed in an infected cell binds at the plasma membrane to ACE2 on another cell. This requires S priming by TMPRSS2 or ADAM10. The priming step may happen in trans, while SARS-CoV-2 is still bound to ACE2 or after insertion of the S2 subunit—which contains the fusion peptide—into the membrane of the ACE2-expressing cell, which may be seen as a cleavage in cis. Inhibitors of TMPRSS2 (e.g., camostat) or ADAM10 reduce cell fusion and syncytia formation. The figure was created with BioRender.
Model for the role of ADAM proteases in SARS-CoV-2 viral entry and lung cell syncytia formation A, B. The SARS-CoV-2 spike protein S consists of the two subunits S1 and S2 and binds through S1 to ACE2 on host cells, which is required for viral fusion with host cells (A) and for syncytia formation (B). (A) TMPRSS2 may cleave S in trans and allow for the fusion of SARS-CoV-2 with the host cell plasma membrane. In contrast, ADAM17 may either cleave S in trans at the plasma membrane or may cleave it once the fusion peptide of S has inserted into the host plasma membrane after endocytosis, which is conceptually similar to a cleavage in cis. After ADAM17 cleavage, the fusion of SARS-CoV-2 with the host cell is likely to happen after endocytosis. Inhibition of ADAM17 reduces viral uptake and fusion. (B) For syncytia formation, the S protein expressed in an infected cell binds at the plasma membrane to ACE2 on another cell. This requires S priming by TMPRSS2 or ADAM10. The priming step may happen in trans, while SARS-CoV-2 is still bound to ACE2 or after insertion of the S2 subunit—which contains the fusion peptide—into the membrane of the ACE2-expressing cell, which may be seen as a cleavage in cis. Inhibitors of TMPRSS2 (e.g., camostat) or ADAM10 reduce cell fusion and syncytia formation. The figure was created with BioRender.
This study identified ADAM17 as a host factor involved in SARS-CoV-2 infection in the lungs. Additionally, the authors found ADAM10 to be an important component of lung cell syncytia formation, an important pathological marker for patients with COVID-19.
A previous study identified ADAM10 and ADAM17 as metalloproteases that are mainly involved in cell communication. For instance, ADAM10 plays an important role in Notch signaling, whereas ADAM17 is involved with intestinal barrier formation and maintenance. Scientists revealed that both the proteases are connected with pathophysiological conditions, such as inflammation (ADAM17) and Alzheimer's disease (ADAM10). Several studies have reported that both ADAM10 and ADAM17 shed numerous membrane protein substrates.
In this study, researchers used genetic ablation and pharmacological inhibition to identify the new role of ADAM17 in SARS-CoV-2 pathogenesis. They found metalloprotease inhibitors could effectively block ADAM17 and, thereby, reduce SARS-CoV-2 infection in the lung cells of humans. Scientists also demonstrated pathogenesis, which typically develops at a later stage of SARS-CoV-2 infection, i.e., syncytia formation of lung cells. This development might have occurred due to the fusion of infected cells expressing S protein and containing ACE2.
The authors of this study stated that even though ADAM10 and ADAM17 function at different phases of SARS-CoV-2 infection, both proteases function via proteolytic priming of the S protein. Additionally, scientists demonstrated that ADAM10 and ADAM17 could prime at or near to S2 site. This could be because both these proteases convert the S2 domain of S protein in vitro to a similarly-sized S2 fragment, analogous to TMPRSS2.
In this study, researchers confirmed that different proteases, such as TMPRSS2, ADAM10, ADAM17, and many others, may prime S2 and mediate infection via the fusion of the SARS-CoV-2 virus with host cells and the formation of syncytia in S-expressing SARS-CoV-2-infected cells with host cells.
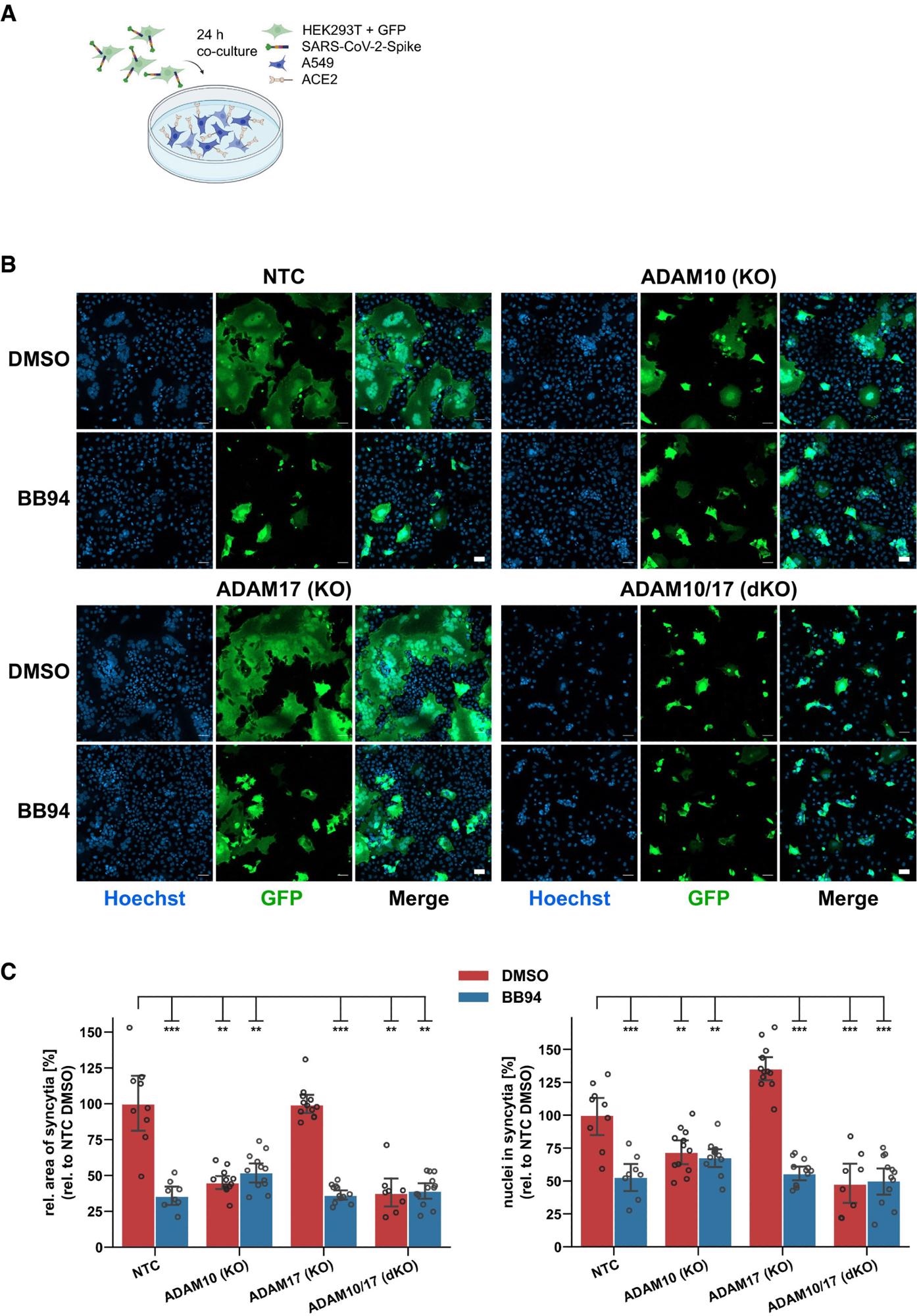
ADAM10 is required for lung cell syncytia formation. Experimental scheme of syncytia formation assay. Donor cells (HEK293T) co-expressing the SARS-CoV-2 spike protein along with GFP were co-cultured with A549-ACE2 NTC or KO acceptor cells. After 24 h, co-cultures were analyzed by confocal microscopy. Representative images of syncytia formation assay. Large syncytia were observed for NTC and ADAM17 KO cells but were strongly reduced when ADAM10 was knocked out, either alone or together with ADAM17 or upon inhibition with BB94 (10 µM). For both ADAMs, the cell lines obtained with gRNA sequence 1 were used. Images show GFP fluorescence and Hoechst staining. Scale bars, 50 µm. Quantification of images from fluorescence microscopy (B). Left plot: the area of the fused cells in green (GFP) was quantified and normalized to the Hoechst signal (blue) of the entire image to account for differences in cell density. Right plot: The nuclei within syncytia were determined by calculating the ratio between the Hoechst signal within syncytia and the total Hoechst signal in the entire image. The data are normalized to NTC DMSO (N ≥ 8). Two-way ANOVA with Tukey’s correction for multiple comparisons. All data are represented as mean ± 95% CI of at least three independent experiments. **P < 0.01, ***P < 0.001. See also Fig EV4.
As A549-ACE2 cells cannot express TMPRSS2 and the rate of infection is drastically reduced upon administration of lysosomal cathepsin inhibitor and ADAM17 inhibitor, researchers presumed that ADAM17 functions in the lysosomal entry pathway, either before endocytosis or after endocytosis, at the plasma membrane. Interestingly, a fusion of cells during syncytia formation occurs at the plasma membrane, which depends on ADAM10 in the TMRPSS2-negative A549-ACE2 cells.
Conclusion
The authors of this study established that both ADAM17 and ADAM10 contribute to COVID-19 pathogenesis. These proteases can independently affect the entry of SARS-CoV-2 into the host and pathological lung cell fusion. Scientists stated that mechanistically, both the above-mentioned proteases cleave SARS-CoV-2 spike protein in vitro. The current study revealed that ADAM10 and ADAM17 could be potential drug targets to inhibit SARS-CoV-2 invasion into the host cell and lung cell syncytia formation.
Journal reference:
- Jocher, G. et al. (2022) ADAM10 and ADAM17 promote SARS-CoV-2 cell entry and spike protein-mediated lung cell fusion. EMBO Reports. https://doi.org/10.15252/embr.202154305, https://www.embopress.org/doi/full/10.15252/embr.202154305