Despite what is known thus far, coronavirus disease 2019 (COVID-19), caused by a member of the Coronaviridae family, SARS-CoV-2, persists as a pandemic that needs immediate healthcare intervention. COVID-19 adversely affects both public and private endeavors, leaving hardship, morbidity, and destruction in its wake. The high morbidity and mortality associated with COVID-19 and its pandemic version are linked significantly with the SARS-CoV-2 transmission rate from individual to individual.
COVID-19 management with antiviral medications and vaccines has proven complicated due to the constant mutations of SARS-CoV-2, with new viral variants arising frequently. As a result, effective, novel, and more efficacious medications against coronaviruses are required.
About the study
In the present work, the scientists explored potential host miRNAs that could be used as broad-spectrum antiviral medicines resistant to coronaviruses. They particularly emphasized the following coronaviruses, Middle East respiratory syndrome CoV (MERS-CoV), bovine CoV (BCoV), SARS-CoV-2, and SARS-CoV. miRNAs are tiny molecules that bind to messenger RNA (mRNA) and regulate translation and transcription.
Further, RNA-dependent RNA polymerase (RdRp) found in coronaviruses is an essential enzyme for the virus's life cycle. Therefore, the researchers hypothesized that host miRNAs could impede coronavirus replication by interacting with RdRp mRNA. To find out this, they retrieved the nucleotide sequences of open reading frame 1ab (ORF 1ab) and utilized them to search miRNA databases interrogating miRNAs that can attach them. The team used multiple bioinformatics methods to anticipate and discover the most influential host miRNAs.
The analytical pipeline used in this research, from sequence curation to interactome circuits, was a slight revision of the authors' previously published model. First, the investigators used https://ngphylogeny.fr/ to create a phylogenetic tree to assess the evolutionary pattern and proximity between the RdRp domain and the 13 coronaviruses. Next, they chose four frequent coronaviruses in cattle and humans (SARS-CoV, SARS-CoV-2, BCoV, and MERS-CoV) for additional investigation to see if host cellular miRNA may address CoV RdRp.
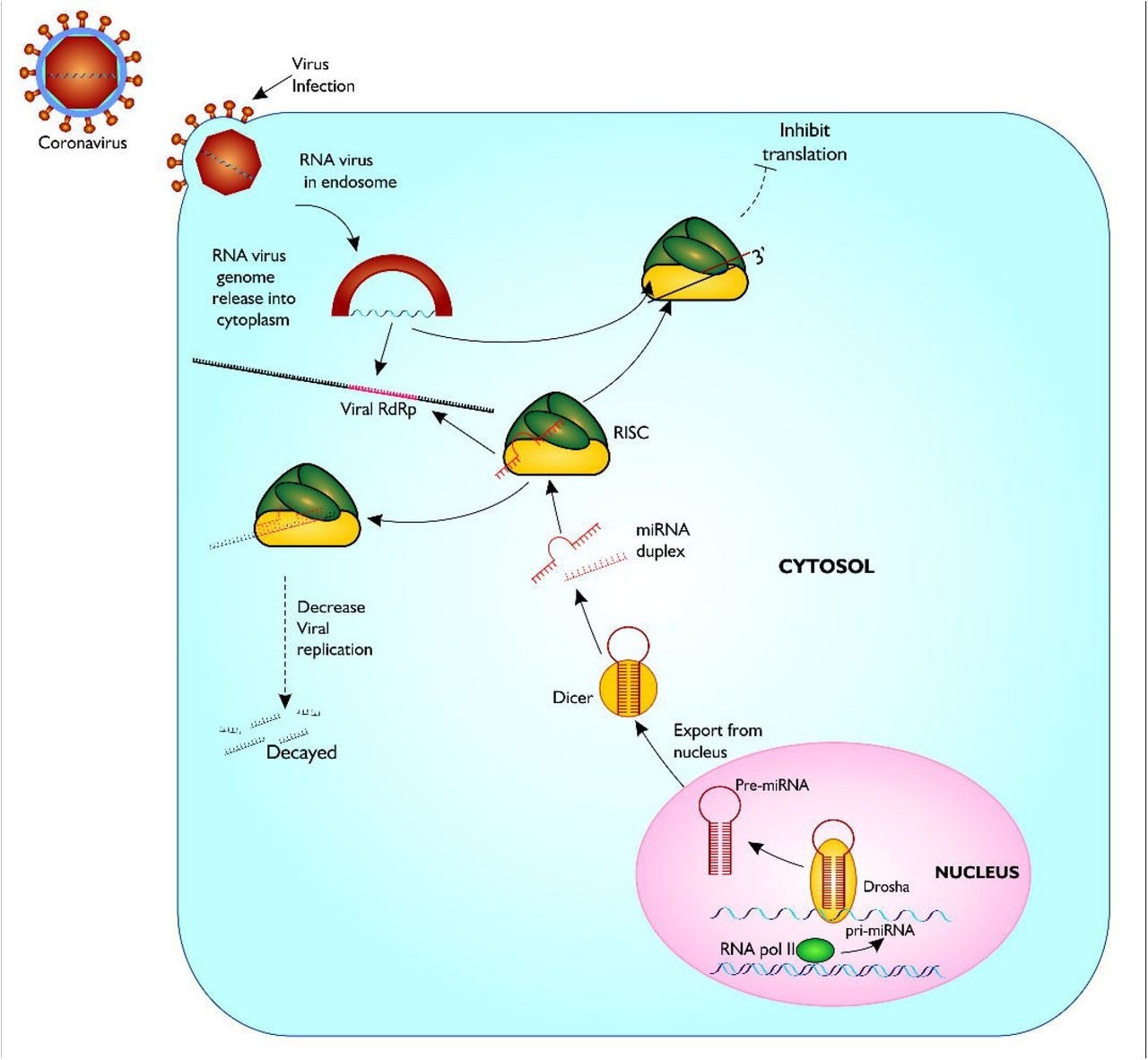
Proposed model of miRNA biogenesis and base pairing with coronavirus RdRp mRNA sequence. The figure gives a description of coronavirus infection of host, and release of host miRNA to base pair and degrade the virus or inhibit translation. Figure created with sketch pad.
Results
Using evolutionary analyses, the researchers discovered that the miRNA binding region across the RdRp sequence of numerous coronaviruses was conserved. Interestingly, the RdRp sequence for MERS-CoV and βCoV England 1 showed no change in the pairwise distance finding, suggesting they probably have the same attaching domain for host miRNAs. The RdRp gene's genetic conservation across various viruses indicates a high positive selection for this area. Additionally, it validates that the enzyme coded by this area was required for the replication of practically all RNA viruses, bolstering its suitability as a target for miRNA drugs.
The investigators discovered 27 miRNAs that address RdRp mRNA of different CoVs, three of which, hsa-miR-579-3p, hsa-miR-664b-3p, and hsa-miR-1283, target SARS-CoV-2, BCoV, and SARS-CoV. Besides, bta-miR-374c, bta-miR-374a, and bta-miR-374b were three bovine miRNA homologs of hsa-miR-374a-5p.
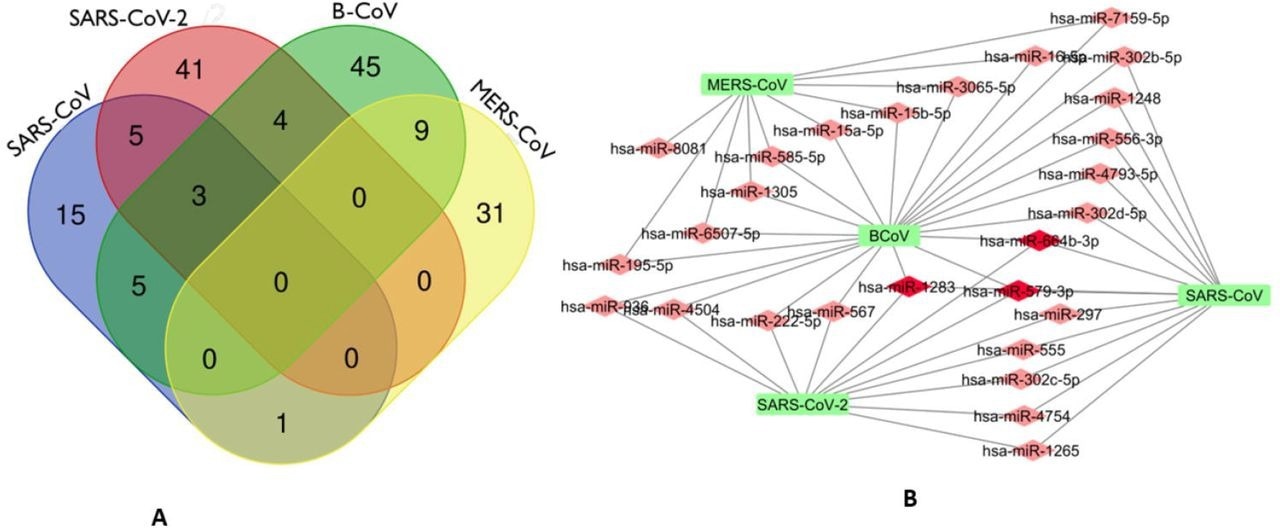
Venn Diagram (http://bioinformatics.psb.ugent.be/webtools/Venn/) showing the number of predicted human miRNA that can target multiple coronaviruses. The number in the intersection/overlapping regions represent the number of miRNAs that can concomitantly target the coronaviruses represented by the intersected shape. (a). Network connections among miRNAs and RdRp of SARS_CoV, B_CoV, MERS_CoV and SARS_CoV-2. B. Generated using Cytoscape 3.7.2
Some miRNAs discovered possessed numerous binding locations within the RdRp domain, improving the chances of binding and lowering off-target impacts, enhancing their potential to become antiviral drug options. Administration or upregulation of hsa-miR-579-3p, hsa-miR-664b-3p, or hsa-miR-1283 may play a dual function in limiting viral degradation/replication and inhibiting cancer progression, according to the scientists, because they have previously shown to have onco-protective properties.
Collectively, the current research sheds light on a potential alternative pathway for targeting and limiting CoV replication through host non-coding RNA, i.e., miRNAs, that target RdRp enzymatic activity instead of the commonly used anti-coronavirus medicines. miRNAs have the potential to be a broad-range antiviral, with hsa-miR-579-3p, hsa-miR-664b-3p, and hsa-miR-1283 showing great promise.
Conclusions
According to the authors, non-coding RNA, such as miRNAs, has not been investigated as an antiviral medication alternative.
The team used numerous computational methods in the current research to investigate genome plasticity and identify potent host miRNAs that can attach to coronaviruses' RdRp sequence area. Even though the viral genome was known to be diverse, they showed that the RdRp sequence was highly conserved across different coronavirus species, implying evolutionary preference and hence a plausible signal for genome addressing.
The present study provides an alternate COVID-19 therapy strategy that involves limiting the translation of the most critical protein (RdRp) for SARS-CoV-2 replication via host miRNAs, resulting in decreased viral pathogenicity and propagation. Given the importance of RdRp in viral survival and multiplication, the researchers discovered host miRNAs attaching to RdRp mRNA in four coronaviruses, leading to its disintegration, thus regulating RNA virus replication and pathogenesis and creating a new avenue for CoV therapeutic targets. Particularly promising were hsa-miR-664b-3p, hsa-miR-1283, hsa-miR-579-3p, and hsa-miR-374a-5p, harboring bovine homologs bta-miR-374b, bta-miR-374c, and bta-miR-374a.
In summary, the present work paves the way for establishing non-coding RNAs as a wide-spectrum antiviral medication. Moreover, it offers the groundwork for subsequent research to confirm the successful binding of discovered miRNAs to coronavirus RdRp sequences in vitro or in vivo.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Deciphering inhibitory mechanism of coronavirus replication through host miRNAs-RNA-dependent RNA polymerase (RdRp) interactome; Olanrewaju B Morenikeji, Muyiwa S Adegbaju, Olayinka S Okho, Asegunloluwa E Babalola, Anastasia Grytsay, Olubumi A Braimah, Mabel O Akinyemi, Bolaji N Thomas. bioRxiv preprint 2022, DOI: https://doi.org/10.1101/2022.06.18.496304, https://www.biorxiv.org/content/10.1101/2022.06.18.496304v1
- Peer reviewed and published scientific report.
Morenikeji, Olanrewaju B., Muyiwa S. Adegbaju, Olayinka S. Okoh, Asegunloluwa E. Babalola, Anastasia Grytsay, Olubumi A. Braimah, Mabel O. Akinyemi, and Bolaji N. Thomas. 2022. “Deciphering Inhibitory Mechanism of Coronavirus Replication through Host MiRNAs-RNA-Dependent RNA Polymerase Interactome.” Frontiers in Genetics 13 (August). https://doi.org/10.3389/fgene.2022.973252. https://www.frontiersin.org/articles/10.3389/fgene.2022.973252.