The cells of the body communicate with each other in a plethora of ways. The C- type Lectin Receptors (CLRs) is one such means of signaling, belonging to the category of pathogen recognition receptors (PRR). These are useful when innate immune cells called antigen presentation cells (APCs) need to pick up pathogen-associated molecular patterns (PAMPs), and when the normal immune response needs to be activated. There is a broad range of CLRs depending on the cell type, including Dectin-2, DC-SIGN, and Langerin.
When CLRs interact with their carbohydrate ligands on dendritic cells (DCs), one important type of APC, the immune response is either activated or modulated to allow tolerance. In particular, this is done through antigen presentation in lymphoid organs, and secondarily via cytokine release. DCs are found at sites where pathogens first come into contact with the body, such as epithelial surfaces of the upper respiratory tract and the lung alveoli.
One mechanism of immune evasion or even harnessing immune activity for the benefit of the pathogen is to bind to CLRs on the surface of the DCs in order to facilitate their transfer towards their real host receptor and cell entry. CLRs, especially L-SIGN (or DC-SIGNR) or DC-SIGN, are well-known for this role in the entry of viruses like HIV, cytomegalovirus, dengue, Ebola, and Zika virus.
HIV, for instance, binds to DC-SIGN, which allows the DC to be directly infected but also passes on the bound virus to T cells, the actual target cell for this virus. This is called cis and trans infection. These receptors also enhance the infection of host cells by SARS-CoV-1.
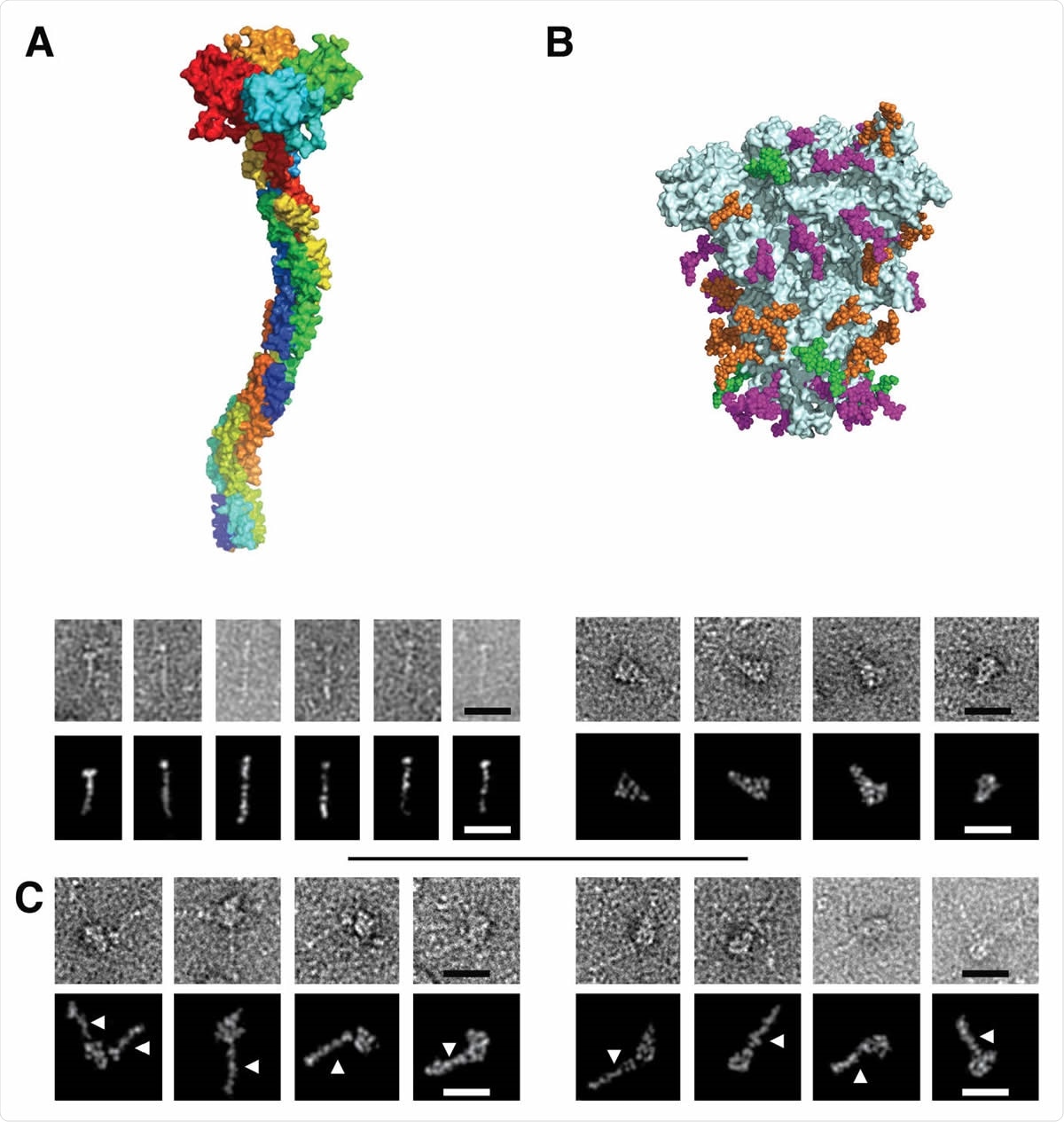
Figure 3. Electron Microscopy micrographs of DC-SIGN/S protein complexes (A) DC-SIGN. Top: model of DC-SIGN ECD tetramer adapted from Tabarani et al (2009). On the bottom: Negative staining images of DC-SIGN. Top row: original images; bottom row: Photoshop processed images. The scale bar represents 25 nm. (B) Spike protein. Top: model of the glycosylated Spike adapted from model of Casalino et al pdb 6vsb). Glycan sites are represented with color code derived from the work of Crispin et coll. (Watanabe et al., 2020a), according to oligomannose-type glycan content, in green (80-100%), orange (30-79%) and magenta (0-29%). On the bottom: Negative staining images of spike protein. Top row: original images, bottom row: Photoshop processed images. The scale bar represents 25 nm. (C) Complex between DC-SIGN and spike protein. Negative staining image of the complexes between DC-SIGN and spike protein. The white arrows highlight DC-SIGN molecules. Top row: original images; bottom row: Photoshop processed images. The scale bar represents 25 nm.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
SARS-CoV-2 Spike Protein
Coronaviruses are characterized by the presence of numerous spikes, with the surface of each spike head and stalk being covered by glycans to the tune of 60% to 90%, respectively. The glycans are mostly conserved between the two SARS-CoVs, in chemical composition, and in position. This has been suggested to create a glycan shield against neutralizing antibodies. Earlier studies using molecular dynamic simulations have also indicated possible mechanisms via the spike glycans can modulate ACE2-virus interactions by stabilizing the receptor-binding domain (RBD) in the up conformation.
Another possible method of interaction is that the spike sugars may anchor the virus to CLRs on the host cells since almost 30% are oligomannose sugars that are ligands for CLRs. Some mutations that affect the virulence of the protein are known to affect the level of glycosylation of the spike.
The current study focused on the interactions between the virus and these receptors. The researchers found that CLRs bind multiple sites on the S protein, and this initial adhesion to the host cells is essential for efficient capture, the concentration of virus particles on the host cell surface, and then searching for and attaching to the ACE2 receptor.
The researchers explain, “Although such additional receptors may not promote any fusion step, they can drive viral internalization through endocytic processes or simply by viral adhesion to the host cell, accumulation of viral particles on the cell surface and finally engagement with the primary receptor followed by the fusion event.”
APCs and Pseudovirions
The study went on to examine the role of the CLRs DS-DIGN and L-SIGN in SARS-CoV-2 pseudovirion infection. The host cell used was monocyte-derived DCs (MDDCs) and M2 monocyte-derived macrophages (M2-MDM), which express DC-SIGN. They found that despite the efficient infection of these cells by the pseudovirions, the binding was not via these CLRs since it was not blocked by anti-DC-SIGN antibodies.
However, when the pseudovirions were incubated with MDDCs and then placed on Vero cells, efficient trans-infection took place. This was significantly reduced by anti-DC-SIGN antibodies.
Next, they tested the binding of SARS-CoV-2 to T cells lacking ACE2 receptors and thus incapable of being directly infected. They found that when the pseudovirus was incubated with a mixture of these cells, both those which expressed and those which did not express the CLRs, with Vero cells, they found that the latter were efficiently transfected in the presence of the T cells with CLR expression. Again, this was significantly reduced by anti-DC-SIGN antibodies by 87% and 79%, respectively.
Finally, the researchers used a known sugar that mimics the natural ligand of DC-SIGN. This compound, called PM26, inhibited the binding of DC-SIGN to the spike protein. It also prevented 99% and 77% of the transfection of Vero cells when incubated with a mixture of pseudovirions and pre-treated DCs.
Micromolar Affinity
The binding between these CLRs and the spike protein is similar to a “Velcro effect,” with a multitude of binding sites rather than just one. However, L-SIGN and DC-SIGN both have micromolar affinities for the spike protein, which will result in a stable surface affinity that is higher by several orders of magnitude at the cell surface. The microdomain organization of CLRs allows multiple attachment points to be presented for viral capture.
Accordingly, DC-SIGN enhanced infection by SARS-CoV-2 pseudovirions significantly when the latter were incubated with cells bearing the CLR and then on ACE2-carrying Vero cells.
The study sums up: “Our work shows that DC/L-SIGN are important enhancers of infection mediated by the S protein of SARS-CoV-2 that greatly facilitate viral transmission to susceptible cells.” While DC-SIGN is found in immature DCs in the submucosa and in the tissue-resident macrophages, including the alveolar macrophages, L-SIGN is found in human type 2 alveolar cells and lung endothelium.
Implications
The expression of CLRs like the above is modulated by IFN, TGF-β, and other anti-inflammatory molecules. However, when ligand binding occurs at the DC-SIGN or L-SIGN receptor, an immune response occurs with the release of pro-inflammatory cytokines like IL-6. This has been found to contribute to the cytokine storm seen in severe COVID-19 with its often fatal termination. SARS-CoV-2 can also prevent the production of type I and type III interferons and thus upregulate the expression of these CLRs.
This study points to the importance of developing DC-SIGN and L-SIGN antagonists to reduce the severity of infection by inhibiting their activity or expression, just as PM26 does. This can not only bind the lectin but also enhance its internalization, reducing the number of CLRs available for the virus to bind it and transfect susceptible cells. This could also induce a pro-inflammatory response on binding to DC-SIGN, which could help reduce the severity of the infection. Thus, this is a promising area of research.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources