Researchers in the United States have presented computational structural models of the binding interactions between coronaviruses and host cells that could be used in the future to screen for strains that are likely to jump from animals to humans.
ACE2 is the main target receptor that the viral spike protein found on severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) binds to and fuses with, in order to gain entry to host cells.
The team used comparative computational analysis of bound and unbound virus in the models to establish which key residues are involved in the SARS-CoV-2 RBD – ACE2 binding interaction and to develop a model of RBD evolution that could serve as a conceptual framework for future studies.
The researchers say the binding model could improve the interpretability of findings regarding the evolutionary diversity of coronavirus strains that infect reservoir species such as bats.
A pre-print version of the article is available on the server bioRxiv*, while the article undergoes peer review.
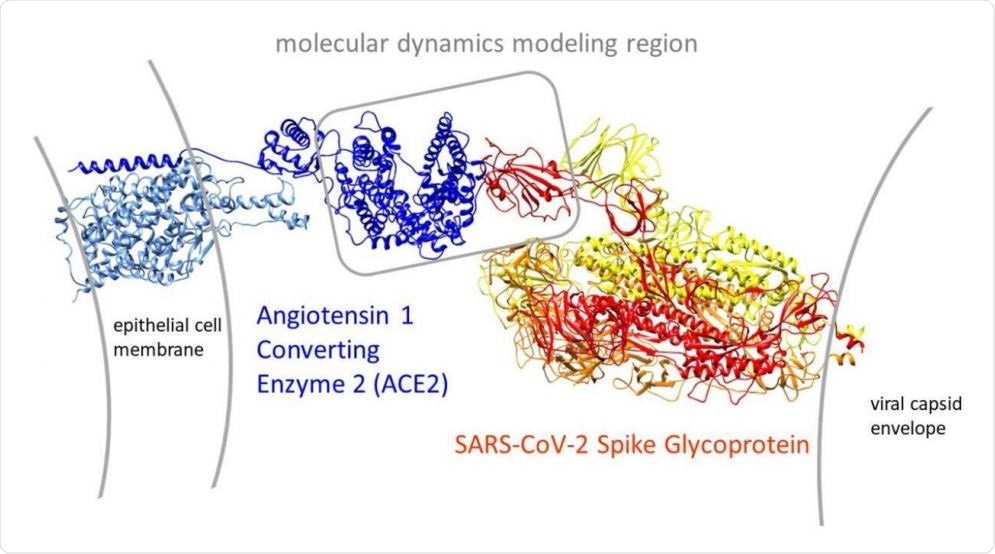
Overview of the open conformation SARS-CoV-2 spike glycoprotein (PDB: 6vyb) interaction with its human target cell receptor, angiotensin 1 converting enzyme 2 (ACE2 PDB: 6m17). The gray box in the center shows our modeling region that bounds most of the subsequent molecular dynamics simulations in the study.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Understanding the similarities between SARS-CoV-2 and other coronaviruses
The unprecedented infectivity of SARS-CoV-2, the agent that causes coronavirus disease 2019 (COVID-19), has led researchers to question the structural, functional and evolutionary similarities it shares with other coronaviruses that have caused disease outbreaks.
One of the main focuses in comparative studies of infectivity is the viral spike protein, given that its interaction with the target receptor ACE2 is the initial step that enables host cell entry.
The spike protein comprises two functional subunits. The S1 subunit contains the RBD that interacts directly with ACE2, and the S2 subunit contains the fusion machinery that enables the virus to fuse to the host cell membrane.
The boundary between these two subunits forms a loop that is exposed in the human coronaviruses OC43, HKU1, MERS-CoV, and SARS-CoV-2.
However, this loop is not exposed in SARS-CoV-1, the coronavirus that caused the 2002 to 2003 SARS outbreak.
Patrick Rynkiewicz (Rochester Institute of Technology) and colleagues say this points to a complex evolutionary landscape for SARS-CoV-related coronaviruses that cannot be fully understood using only amino acid or nucleotide sequence comparisons.
The team says that understanding the sequence and structural variations that affect the molecular dynamics of ACE2-RBD binding could help to better inform the design and development of therapeutic agents targeting this interaction.
“The comparative dynamic modeling of present, past, and potential progenitor viral strain interactions to ACE2 and other known targets of past outbreaks can also potentially illuminate critical functional aspects that facilitate zoonotic [animal to human] spillovers involving the betacoronaviruses,” writes the team.
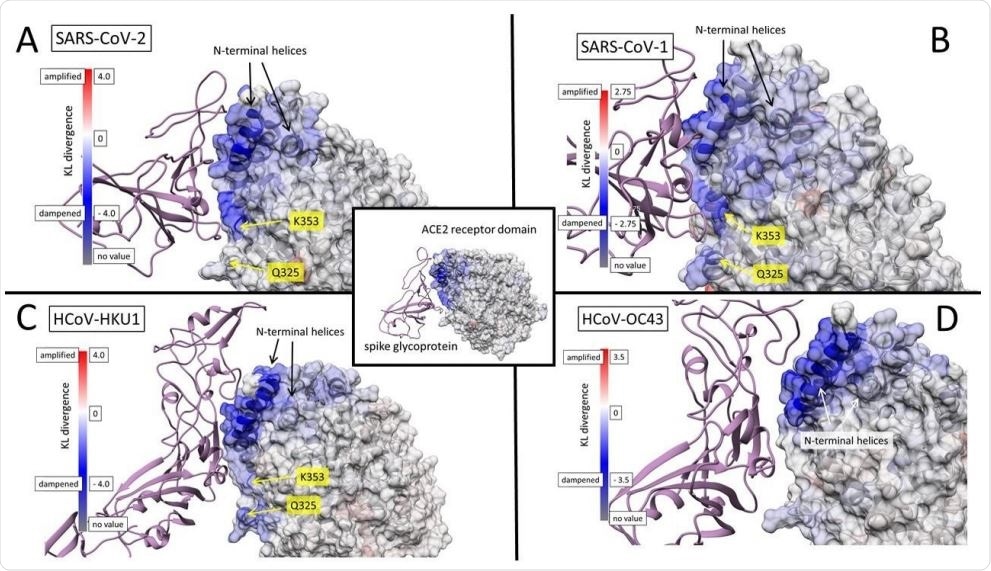
Color maps of dampened atom fluctuations in human ACE2 receptor protein due to the binding of betacoronavirus spike glycoprotein. Shift in atom fluctuations were calculated as the signed symmetric Kullback-Leibler divergence (i.e. relative entropy) of distributions the root mean square time deviations (rmsf or FLUX) for residue averaged protein backbone atoms (i.e. N, Cα, C and O) for ACE2 in its spike bound versus unbound dynamic state. The (A) SARS8 CoV-2 (COVID19) interaction model with ACE2 model (PDB: 6m17) was used as a structural alignment template to model the potential interactions of other human betacoronavirus spike glycoproteins (B) SARS CoV-1 from PDB: 6acg (C) HCoV-HKU1 from PDB: 5gnb, and (D) HCoV11 OC43 from PDB:6ohw. After 1ns of heating and equilibration, the molecular dynamics for the viral bound and unbound states were each sampled over 200 production runs at 0.2ns in charge neutralized explicit solvent periodic boundary conditions at temperature of 300K and pressure of 1 atmosphere. Note: blue mapping indicates dampening of atom motion while red indicates amplification of motion.
What did the study involve?
Now, Rynkiewicz and the team have used a combined methodology and software tool to analyze and visualize comparative protein dynamics and hybrid models of known betacoronvirus spike target interactions.
The team compared the motions that characterize the interaction between SARS-CoV-2 Spike and ACE2 with similar interactions seen with other coronaviruses. The researchers also tested whether the presence of a widely used blood pressure drug might alter the binding recognition of the SARS-CoV-2 spike due to the stabilizing effect it has on ACE receptors.
Statistical comparisons of molecular dynamics identified key residues involved in the spike RBD-ACE2 binding that occurs during infection with SARS-CoV-2 and other coronaviruses.
The ACE2 residues involved are K353, Q325, and 386-AAQPFLL-392.
In addition, the prevalence of ACE2 N-terminal helix interactions among all of the viral strain models studied led the team to think that this is a necessary prerequisite interaction for all zoonotic spillovers that involve coronaviruses.
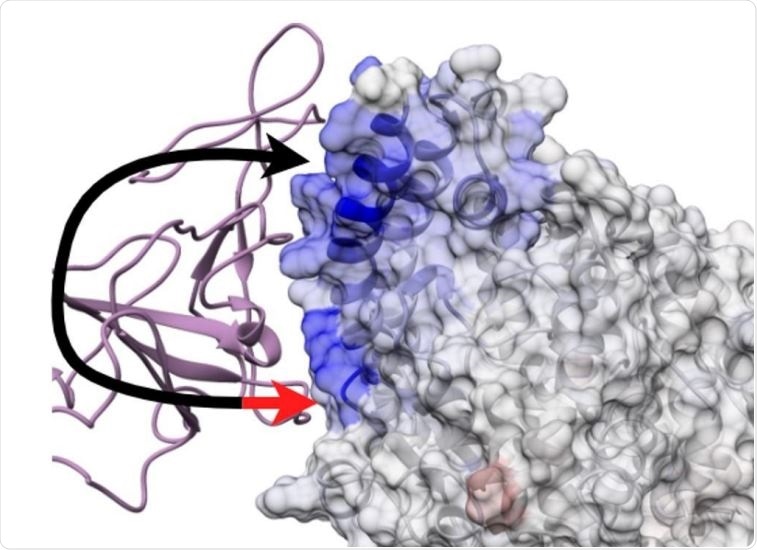
Two touch model of protein interaction. Black arrowhead - a promiscuous viral site of rapid dynamic interaction common across all outbreaks and also present in potential zoonotic encounters. Red arrowhead - a site active in more pathogenic outbreaks characterized by more complex dynamics enabling articulation between novel sites on human ACE2.
Creating models of viral evolution
Next, the team used the findings to create a “one-touch” model of viral evolution to assess the predisposition of viruses of zoonotic origin to species crossover based on an initial interaction with ACE2 N-terminal helices.
This showed that following infection of the human host, an additional anchor point among the K353, Q325, and 386-AAQPFLL-392 residues in ACE2 appears to be established by the RBD.
The team says this results in a “two-touch” model that is seen more specifically in highly pathogenic species of human coronavirus.
Finally, the researchers found that when the hypertension drug lisinopril was bound centrally within the ACE1 and ACE2 structures, the molecular motion within the spike RBD-ACE2 protein complex seemed to stabilize at many locations along the complex interface, which included the two key touchpoints in the proposed model.
Rynkiewicz and colleagues say it should be noted that no clinical conclusions should be drawn from this finding, such as whether this drug should be prescribed or not.
“Rather, it demonstrates the significant effect of external binding molecules on the protein system and provides the impetus for further clinical research on any potential correlation between ACE inhibitor usage and the likelihood or severity of SARS-CoV-2 infection,” they write.
Future directions
The researchers say the key residues they identified in the SARS-CoV-2 RBD-ACE2 binding interaction and the two-touch model of RBD evolution they developed provide a conceptual framework for future functional mutagenesis studies of this system.
“Future surveys of betacoronavirus strains circulating in past, present, and future human populations as well as molecular and clinical investigations of SARS-CoV-2 infection in the presence of other ACE inhibitors like lisinopril will be informed by the model interpretation of binding interaction that we present in this study,” they conclude.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources