Even as the pandemic of COVID-19 progresses over the world, interest is growing in the structure of the virus that causes severe pneumonia cases. While the genome has been sequenced in hundreds of strains from all over the world, the number of mutations in the viral genome has also grown steadily.
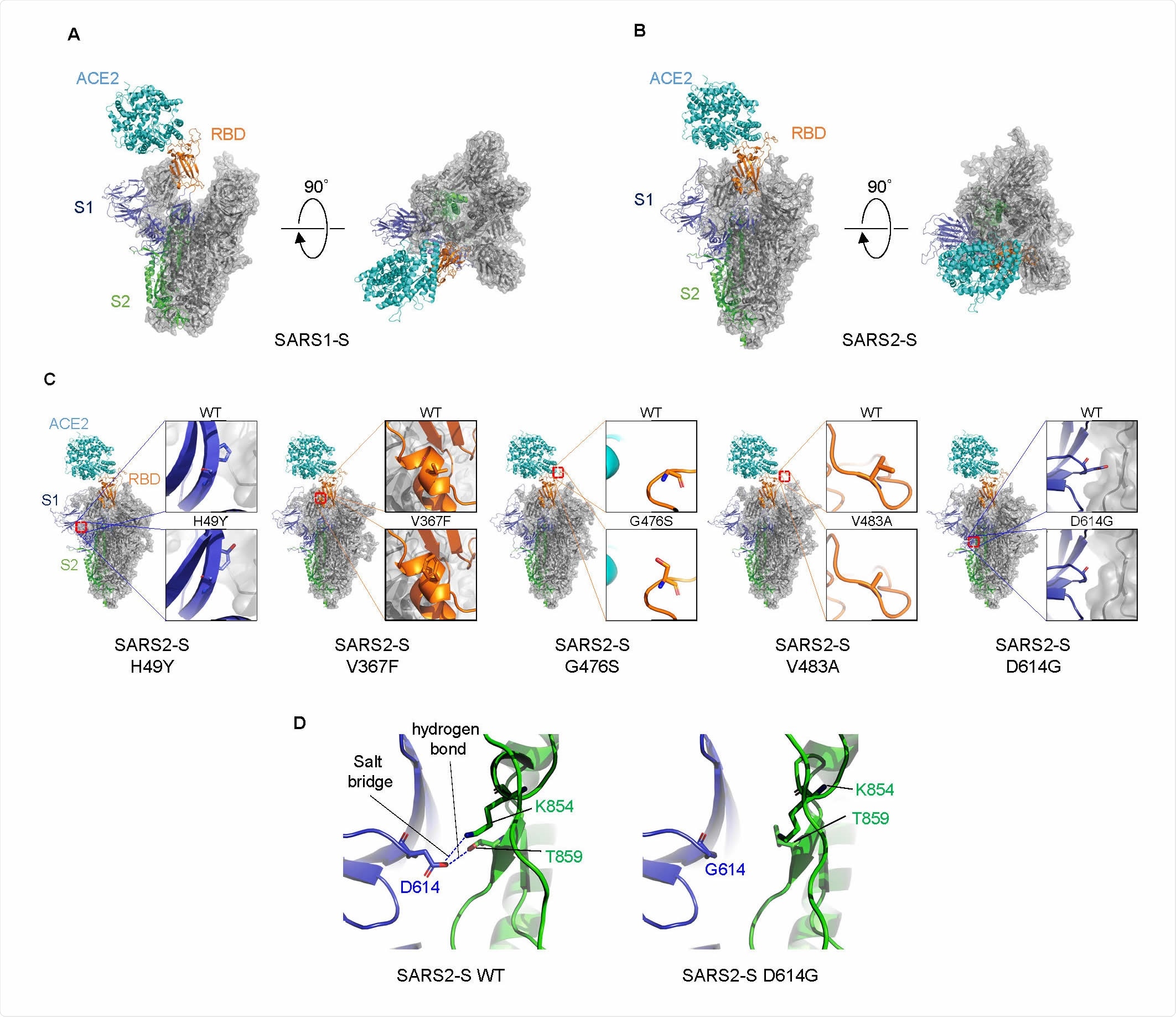
Structural comparison of S proteins.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Mutations in the S Protein
Mutations have been found to occur in all regions of the virus, including the ORF1a, ORF1b, ORF3, ORF8, the nucleocapsid or N protein, and the spike or S protein. However, spike protein mutations are vitally important since they affect the part of the virus that is necessary for the initial step of viral entry, namely, binding to the host human cell receptor, the angiotensin-converting enzyme (ACE2), which triggers cleavage by the transmembrane serine protease 2 (TMPRSS2).
Both the ACE2 receptor and the TMPRSS2 are expressed at high levels in the airway, lung and oronasal epithelium, and also in the intestines. To gain an idea of how much virus enters the cells via S protein activity, which will also show how much inhibitory or enhancing activity each mutation has, the current stud used a new measurement technique.
Quantifying Viral Entry into Host Cells
The technique uses a lentivirus as the vector, carrying a small peptide termed HiBiT that acts as a luminescent tag to precisely account for viral entry, and gauge even small differences which are later amplified over multiple replication cycles. Using this, the researchers first quantified the baseline level of viral entry of lentiviruses pseudotyped with the S protein of either the SARS-CoV or the SARS-CoV-2 (called SARS-S or SARS2-S respectively).
Coexpression of ACE2 and TMPRSS2 Necessary for Viral Entry
They observed efficient viral entry by the SARS-S-pseudovirus into ACE2-expressing cells, but much less efficient entry by the SARS2-S. In other words, ACE2 alone is not enough to promote the entry of SARS-CoV-2. The next step was, therefore, to coexpress ACE2 and TMPRSS2.
They found a marked increase in viral entry by SARS2-S protein, but only a moderate increase in that mediated by the SARS-S. In their words, “SARS2-S-mediated entry into cells is more highly dependent on TMPRSS2 coexpression than that of SARS-CoV, suggesting that they may have different cellular tropisms to some extent.”
S Protein Incompletely Adapted to ACE2
Even so, SARS-S and SARS2-S viruses differed by a factor of 25 in their entry, which led the researchers to think that the latter S variant might possibly be less well incorporated into the lentiviruses relative to the former as a result of poor compatibility. Looking at the amino acid differences in the cytoplasmic tail and transmembrane domain, they generated a mutant SARS2-S containing C1247A, as well as a chimeric form of SARS2-S with the above domains belonging to SARS-S.
At this point, viral entry was comparable with all SARS2-S pseudoviruses, as well as the incorporation of the virion into the cell. This indicates that the reason for the lower rate of cell entry seen earlier with SARS2-S was not, as thought, deficient incorporation but the nature of the S protein in this pseudotype.
What were these differences?
The researchers looked at the ability of the S proteins in these pseudotypes to use either ACE2 or TMPRSS2 at a given level of cell surface. Using cell cultures with a high constant ACE2 expression, while modulating TMPRSS2 levels, and the converse, they observed that pseudotypes expressing SARS2-S needed high coexpression of both TMPRSS2 and ACE2 to infect the cell with the greatest efficiency.
This could mean that this spike protein variant is incompletely adapted to ACE2, using it with decreased efficiency and therefore requiring higher TMPRSS2 expression to infect the host cell. This could even perhaps explain the high rate of asymptomatic and mild infection.
D614G Mutation Enhances Cell Entry
The next step was to look for natural S mutations that might affect viral entry. They found five variants, which were incorporated into plasmids, and examined how they affected cell entry on cells, which expressed both ACE2 and TMPRSS2, relative to the wildtype S protein.
The results showed that natural mutations were associated with variable effects, with three of the five being linked to increased entry into such cells. The natural mutant with the highest cell entry was the D614G, which is also the one that defines the rapidly spreading clade A2a (or clade G).
This clade is remarkable for its ability to quickly take the leading position among all isolates soon after being introduced into a locality. In 15 European countries where this clade has become dominant, the doubling time has dropped by almost half, due to this increase in infectivity.
Structural studies on ACE2-S models showed that the SARS-S trimer presented in an open state, with a larger RBD contact area being exposed for specific interaction with ACE2. This could account for the higher binding of SARS-S RBD to the ACE2 receptor than the SARS2-S. If so, it accounts for the recent finding of a lower binding affinity of the latter with ACE2 than the former.
Structural explanations for a mutation-linked increase in viral entry involve the position of various amino acids, especially the aspartic acid at position 614 (D614), which forms hydrogen bonds and salt bridges with other residues at other locations in the S2 subunit of another protomer. Its replacement with glycine (D614G) increases the flexibility of the space between adjacent protomers, because of the short side chain. This allows a smoother dissociation between the S1 and S2 subunits or makes the whole S trimer structure more flexible, allowing the ACE2 to access the RBD more easily. The result is an increased viral entry with this mutation.
D614G Mutation Does Not Affect Antigenicity
Does the D614G mutation change the antigenic properties of the S protein? This was examined via neutralization assays, comparing the neutralizing sensitivity of the wildtype and mutant proteins when exposed to specific antibody-containing serum. In all cases, the patient serum with anti-SARS-CoV-2 antibodies showed efficient neutralization of both wildtype and mutant S protein.
Implications
In short, say the investigators, “These results indicate that the D614G mutation in the SARS2-S protein maintains neutralization sensitivity to the anti-SARS2-S antibodies.” It will, therefore, be susceptible to any currently developed vaccine.
Moreover, the relative inefficiency of viral cell entry could explain the high rate of asymptomatic spread and, therefore, the progression of a localized infection to become a pandemic. The earlier SARS-CoV had a thousand-fold higher replication potential in cell cultures compared to the current virus, which was associated with almost universally symptomatic infection. In contrast, the continued spread of the SARS-CoV-2 virus among humans allows it to gain mutations that increase its infectivity.
This may represent an area of epidemiological learning: the most dangerous viruses are those with high infective potential and a high asymptomatic infection rate, rather than those which are obviously associated with very high mortality. Further surveillance is mandatory to identify changes in SARS-CoV-2 as it continues to spread.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Ozono, S., et al. (2020). Naturally Mutated Spike Proteins Of SARS-Cov-2 Variants Show Differential Levels of Cell Entry. bioRxiv preprint. doi: https://doi.org/10.1101/2020.06.15.151779. https://www.biorxiv.org/content/10.1101/2020.06.15.151779v1
- Peer reviewed and published scientific report.
Ozono, Seiya, Yanzhao Zhang, Hirotaka Ode, Kaori Sano, Toong Seng Tan, Kazuo Imai, Kazuyasu Miyoshi, et al. 2021. “SARS-CoV-2 D614G Spike Mutation Increases Entry Efficiency with Enhanced ACE2-Binding Affinity.” Nature Communications 12 (1): 848. https://doi.org/10.1038/s41467-021-21118-2. https://www.nature.com/articles/s41467-021-21118-2.