The coronavirus disease 2019 (COVID-19) pandemic has continued to wreak havoc throughout the world. The virus that causes this illness, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), enters the host cell via its spike protein.
A new study by researchers at Stanford University, USA, discusses the effect of specific mutations on the sensitivity of the spike protein to elastase, and on the intramolecular conformational changes. The team's findings have been published on the bioRxiv* preprint server.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Whenever one mutation, or a set of mutations, causes the viral variant to acquire any kind of advantage over the ancestral strain, the new variant – by means of natural selection – becomes dominant. A classic example of this regarding the COVID-19 pandemic is the emergence of D614G, which originated in Europe, but quickly became the dominant strain globally.
This displays a mutation from aspartate to glycine at position 614 on the spike protein. Though this results in higher viral loads, the severity of the syndrome is comparable to that of earlier variants. The mutation does not reduce the susceptibility of the variant to neutralizing antibodies elicited by earlier strains.
The rate of mutation of the SARS-CoV-2 genome is very high, with each amino acid of the over 1,200 which make up the spike protein, for instance, having undergone four mutations, on average, over a year. The constant emergence of new variants makes active monitoring of the pandemic a necessity, especially with respect to their enhanced transmissibility.
The UK and South African variants
The UK saw the rise to dominance of the B.1.1.7 or 20B501Y.V1 within a month of its emergence at the end of September 2020. This spreads more readily than the parent strain, with an increase of 40-70%, and has currently been isolated from over 50 countries. It has not been observed to cause more severe disease, however.
Of the 13 mutations found in this variant, the N501Y is the only one that affects the host receptor – the angiotensin-converting enzyme 2 (ACE2). This mutation is in the receptor-binding domain (RBD) of the viral spike protein. However, it has been observed as far back as April 2020 in Brazil, without increased transmissibility, which indicates that it cannot explain, by itself, this feature of the UK variant.
The 501Y.V2 is another variant dubbed the South African variant. It was first identified in the second week of October 2020, and became dominant there in a month. It has six fixed mutations, namely, N501Y, D614G, and four other mutations. These in combination appear to confer increased infectivity but not virulence on this variant.
The current study focused on identifying the underlying mechanism of higher infectivity, using computational platforms, such as Molecular Operating Environment (MOE) analysis. The results could help detect new therapeutic targets.
The researchers found that the mutations were not associated with any given location or domain of the spike, nor did they lead to exposure of all mutations in either the open or closed conformations. This led the team to postulate that the key to increased infectivity might be an increase in susceptibility of the spike protein to activation by proteases, as additional activating sites were acquired via the new mutations. Moreover, the proteases they sought were thought to be somewhat specific to the entry site, rather than widespread in the host, since these mutations failed to increase the virulence of the illness.
Increased elastase cleavage sites
While several proteases have been suggested as potential candidates, such as TMPRSS2, furin and cathepsin L, only neutrophil elastase is richly found in the human nose, relative to other tissues. Moreover, neutrophils are recruited to the nose when the nasal epithelium is infected by SARS-CoV-2, reaching a tenfold increase in local population numbers, and producing threefold higher levels of elastase. They found that the D614G did indeed bring in an additional elastase-activated site on the spike protein.
They also found two new elastase target sites in the UK variant, of which only one is exposed in the ‘open’ conformation of the RBD. The South African variant contains several new elastase cleavage sites, with two being fixed mutations. The researchers point out that proteolytic cleavage need not be always at the S1/S2 interface, but can be at other sites while still making the spike more ready to expose the RBD.
Reduced intramolecular interactions
Another way in which the new mutations could increase viral infectivity is by decreasing intramolecular interactions in the spike and thus making it more prone to the open conformation. This would promote RBD exposure, thus leading to increased ACE2 binding and viral entry. Some earlier reports suggested that the D614G increased the time spent in the open conformation.
The MOE analysis suggests the potential for the RBD to be in the open conformation with the N501Y mutation, because the asparagine residues at this site in the three spike monomers are at a distance of about 14 Å from each other when the trimer is in the closed conformation. A substitution by tyrosine, which is bulkier, would cause steric hindrance, destabilizing the sites of contact between the monomers in this state. It should be noted that MOE results do not suggest a more unstable closed conformation with tyrosine substitution at 501, compared to the reference sequence.
Another such interaction is because of serine982, which can mediate the closed-open transition. This residue and Thr547 on adjacent monomers of the closed spike form a hydrogen bond, which is abolished if serine is substituted by alanine in 501Y.V1. This change would make the spike more favorable to the open conformation.
The Lys417Asn substitution in the South African variant can also favor the open conformation of the spike, by disrupting the proton-π interaction with the Tyr369 residue on the adjacent monomer that occurs in the closed conformation.
RBD variants
The N501Y mutations of both the 501Y.V1 and 501Y.V2 variants, and the Glu484Lys and Lys417Asn of the 501Y.V2 variant, are at the binding interface of the spike-ACE2 complex, and may increase infectivity by promoting binding.
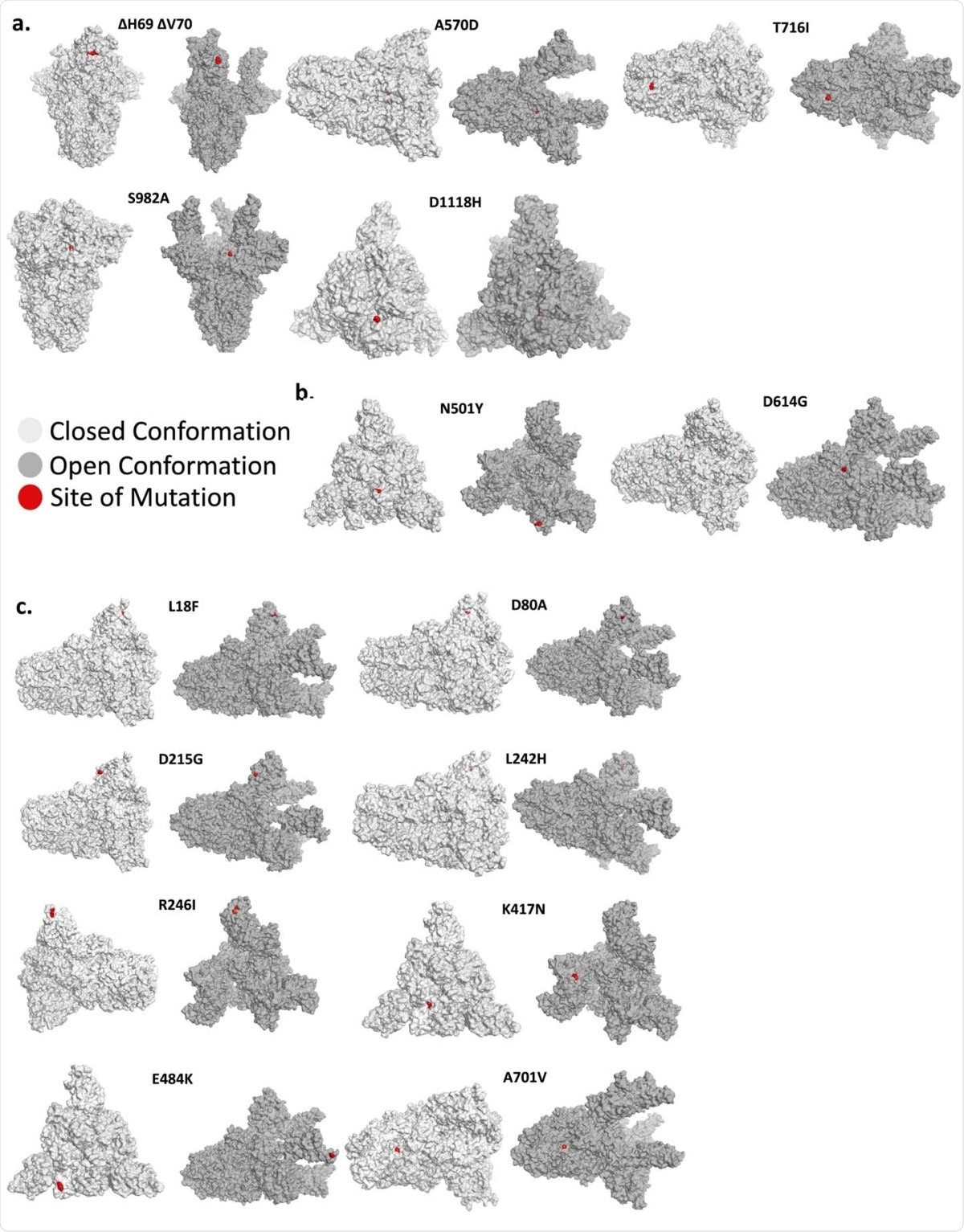
Mutant position in the open and closed conformation of the spike protein. (A) The position of various mutations of the Y501.V1, (B) The N501Y and D614G mutations, which occur in both the Y501.V1 and Y501.V2 variants, and (C) Mutations unique for the 501Y.V2. Each mutation is shown in red in one of the monomers in the 3D structure of the spike protein trimer. Shown is the closed conformation (light gray) and the open (dark gray) conformation.
MOE analysis failed to show any impact on the affinity of binding of the spike RBD to the ACE2 receptor. However, yeast display studies show that when both are found together in a spike variant, the binding affinity is increased twofold and tenfold, for 501Y.V1 and 501Y.V2, respectively. If the mutation affects the RBD, it may also increase the instability of the closed conformation and thus promote binding to ACE2.
The monomers wrap around one another through RBD in the closed trimeric structure and mutations in RBD that abolish these critical inter-monomeric interactions could favor the transition to open conformation.”
This calls for further research on how these mutations affect binding affinity, singly or together, using virions.
What are the implications?
The researchers suggest that by increasing RBD exposure, the mutations also expose a greater number of antigenic determinants per viral particle for binding by neutralizing antibodies. With as many as 80 spike particles per virus, only some need to bind to the host cell ACE2 to accomplish viral entry. On the other hand, activation of more spikes can lead to more exposure of the RBD domain. This will allow increased binding between neutralizing antibodies and the viral particles, agglutinating multiple viruses. This would result in reduced fusion of the viruses to the cell membrane, and thus lower infectivity.
In short, even as the spike protein becomes more easily activated because of the mutations, leading to efficient infection, it simultaneously becomes more susceptible to binding by neutralizing antibodies because of the exposure of its hidden regions.
Elastase is important in promoting viral entry, and several elastase inhibitors have already been approved. These could be tested for their role in preventing SARS-CoV-2 infection, perhaps via intranasal administration. This modeling study should be followed up by experimental proof to gauge the sensitivity of these new variants to existing therapies as well as to develop new targets.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources