With the increasing exploration of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) – the causative pathogen of coronavirus disease 2019 (COVID-19) pandemic – it has become clear that viral particles co-opt human host cell proteins to spread and destroy other host cells. This involves protein-protein interactions of many kinds.
A new study by researchers at Brookhaven National Laboratory and Stony Brook University in the U.S. describes the key interaction between the viral envelope (E) protein and the human cell junction protein, PALS1. The latter is necessary to establish and maintain cell polarity and tight junctions between cells.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The team released their findings on the bioRxiv* preprint server.
Envelope protein mediates viral lifecycle and immune response
The E protein is crucial for the viral lifecycle, being necessary for the curvature of the membrane, so that new viral particles can be assembled. The high levels of E produced during viral replication and transcription are accompanied by its localization to the endoplasmic-reticulum–Golgi intermediate compartment (ERGIC).
The E peptide also provokes host immunity in two different ways. The first is via its transmembrane (TM) pore-forming domain that is associated with the activation of the NLRP3-inflammasome. The TM domain is a pentameric ion channel similar to that of SARS-CoV.
The second is through its binding to the PDZ domain of PALS1 via the PDZ-binding motif (PBM) at its C-terminal domain (CTD). The SARS-CoV-2 E protein has increased binding affinity for the PALS1 PDZ domain compared to SARS-CoV.
E protein mediates virulence
The PDZ domain is found in many proteins, and is central to human immune response regulation, with multiple functions in health and disease. Cell junction proteins containing PDZ domains are often taken over by viral replication processes to increase their virulence.
It seems that the PBM interacts with PALS1, pulling it out from its location in the intercellular junction space to ERGIC, where viral assembly is taking place.
The E protein also interacts with other proteins involved in epithelial junctions. When these are shifted to intracellular locations, devastating consequences set in, including leaking blood vessels, diffuse alveolar damage (DAD), and the cytokine storm.
The outcome is acute respiratory distress syndrome (ARDS), often with a fatal termination in old and/or sick patients with COVID-19.
The PBM contributes to virulence since its absence renders the virus weakened or nonviable. When SARS-CoV with deletions at this site was passaged in the host, the PBM was recovered, suggesting its importance for viral fitness and virulence.
Structural study of PALS1-E complex
The current study aimed to elucidate the structure of this PPI, to help explain how the PALS1 PDZ and SH3 domains are recognized by the E protein PBM.
The PALS1 protein has five domains, namely, two L27 domains at the N-terminal domain and three at the CTD – together called the PSG, which takes part in E recognition. The PSG was modified for this experiment to enhance protein stability.
The PBM-containing 18 amino acid peptide at the CTD of the E protein (Ec18) was synthesized to simplify the examination of complex structure.
Weaker affinity, hydrophobic binding pocket
Using single-particle Cryo-EM (cryogenic electron microscopy), they found that the PALS1-E complex contains one dimeric PSG and a single Ec18 peptide. Of the two PSG components, one had all three domains, but the other did not have a PDZ domain. Instead, a highly disordered region was observed within a class.
The physiological complex formed between the PSG dimer and its Crb-CT ligand is different from the Ec18-PSG complex since in the latter, the PDZ and SH3 domains are rotated about 38° with respect to the GK domain. The former has a stronger binding affinity compared to that of the viral E-PALS1 complex.
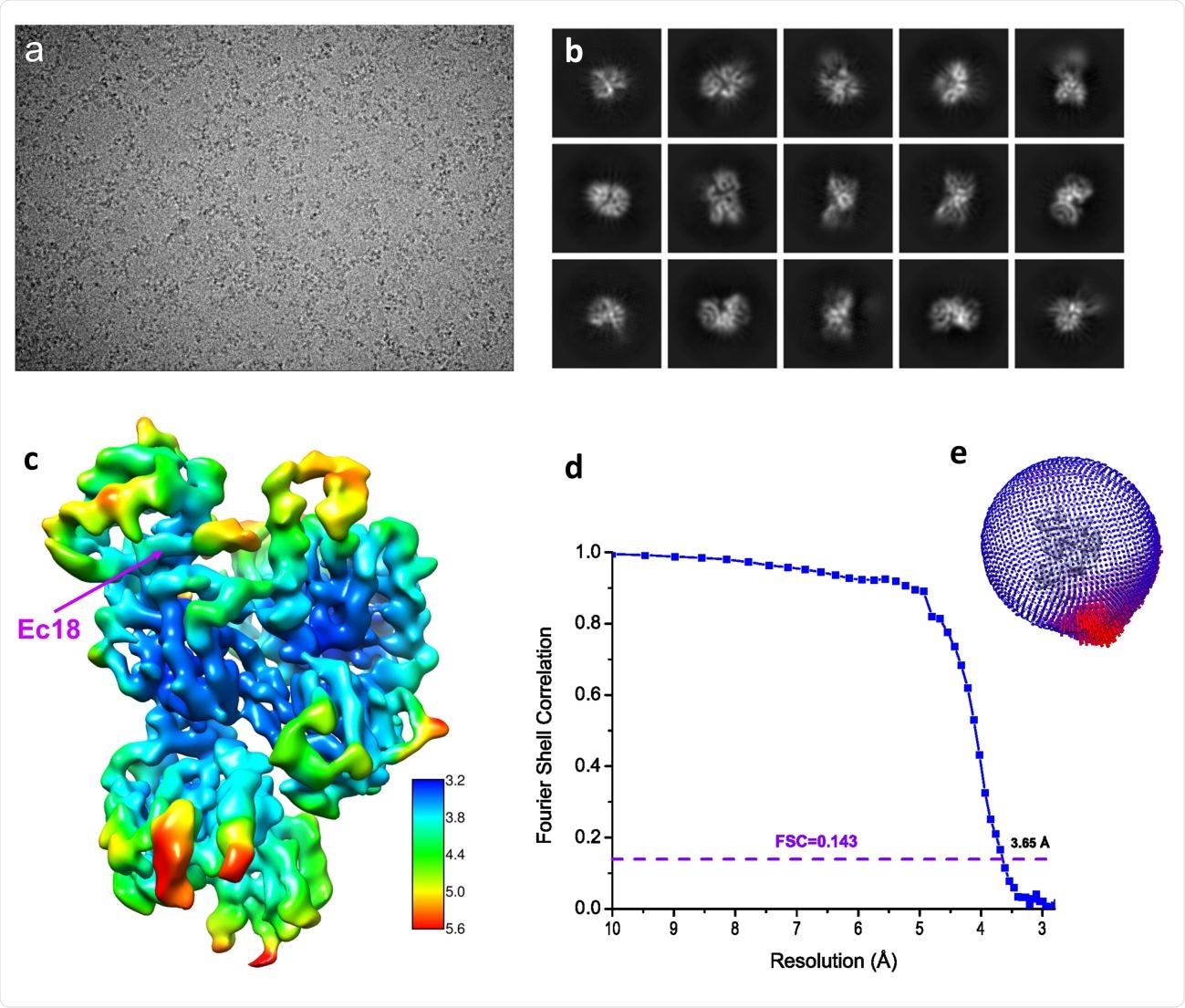
Structure determination by single-particle Cryo-EM. (a) A typical motion-corrected Cryo-EM micrograph. (b) 2D class averages. (c) Reconstructed map colored with local resolutions. (d) Fourier Shell Correlation (FSC) curve for the 3D reconstruction to determine the structure resolution. (e) Orientation distribution for particles used in 3D reconstruction.
Stabilization of binding
The Ec18 peptide was observed to be inserted in a hydrophobic binding pocket, located between the PDZ and SH3 domains, via its binding with the PBM. Phe318 is found to be placed between two hydrophobic amino acids, Leu73 and Val75, which is a prominent feature involved in recognition.
Of the two SH3 and two GK domains in the structure, the former showed quite marked conformational changes accompanying Ec18 binding, involving two leucine residues. These are thought to stabilize the Ec18-PDZ complex.
What are the implications?
The study shows the interactions between the viral E protein and the human PALS1 protein at an atomic level, thus explaining how the envelope mediates viral virulence through its PBM. The viral open reading frame 3 (ORF3) polypeptide could also be a part of this process, involving the co-opting of PDZ-containing proteins to enhance its virulence and fitness.
The structure shows that a peptide inhibitor with a C-terminal isoleucine, leucine or phenylalanine may be able to achieve higher binding affinity with the PALS1 binding pocket because it is inserted more deeply. This reflects the stronger binding between PALS1 and its physiological ligand, with a deeply penetrating isoleucine residue.
An unfavorable arginine in the Crb-CT PBM reduces interactions with a key phenylalanine residue at position 318. Another option could be to substitute this arginine with either leucine or phenylalanine, both of which are hydrophobic.
In the course of the pandemic, many PBM mutations have occurred in the SARS-CoV-2 virus, increasing both virulence and viral fitness. These could be incorporated into a hybrid Crb-CT-viral mutant PBM peptide, which could reduce the interactions between the viral envelope protein and PALS1. This could inhibit virus-induced lung damage and further unfavorable outcomes.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources