The open-source code was obtained by the authors from GitHub and applied to the unglycosylated SARS-CoV-2 spike protein for comparison with glycan conformations obtained by more computationally intense approaches and with experimental data obtained in the wet laboratory.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
How does GlycoSHIELD work?
Most glycan interactions are non-specific and temporary, besides the highly specific and specialized glycan-binding protein domains. Furthermore, the conformation of the glycan is constantly changing based on thermal influences. Therefore, a significant degree of computational effort can be saved by not simulating each individual glycan, instead simulating a smaller subset and grafting them into place on the protein surface, accepting or rejecting the position based on steric hindrance caused by the protein (steric exclusion).
Glycans are capable of influencing the conformation of the protein to which they are bound, and thus the group firstly utilized N-cadherin. This glycoprotein bears five stable domains that are bound by calcium-dependent linkers, wherein in the absence of calcium, the domains are significantly freer. A non-glycosylated model of the protein was prepared, with a traditionally glycosylated version also obtained for comparison to the model having undergone glycosylation by GlycoSHIELD.
A library of 11 high-frequency glycans was generated and applied using GlycoSHIELD. The final glycosylated protein was very similar to the full molecular dynamics simulation (MDS) obtained model in terms of shielding of the protein surface.
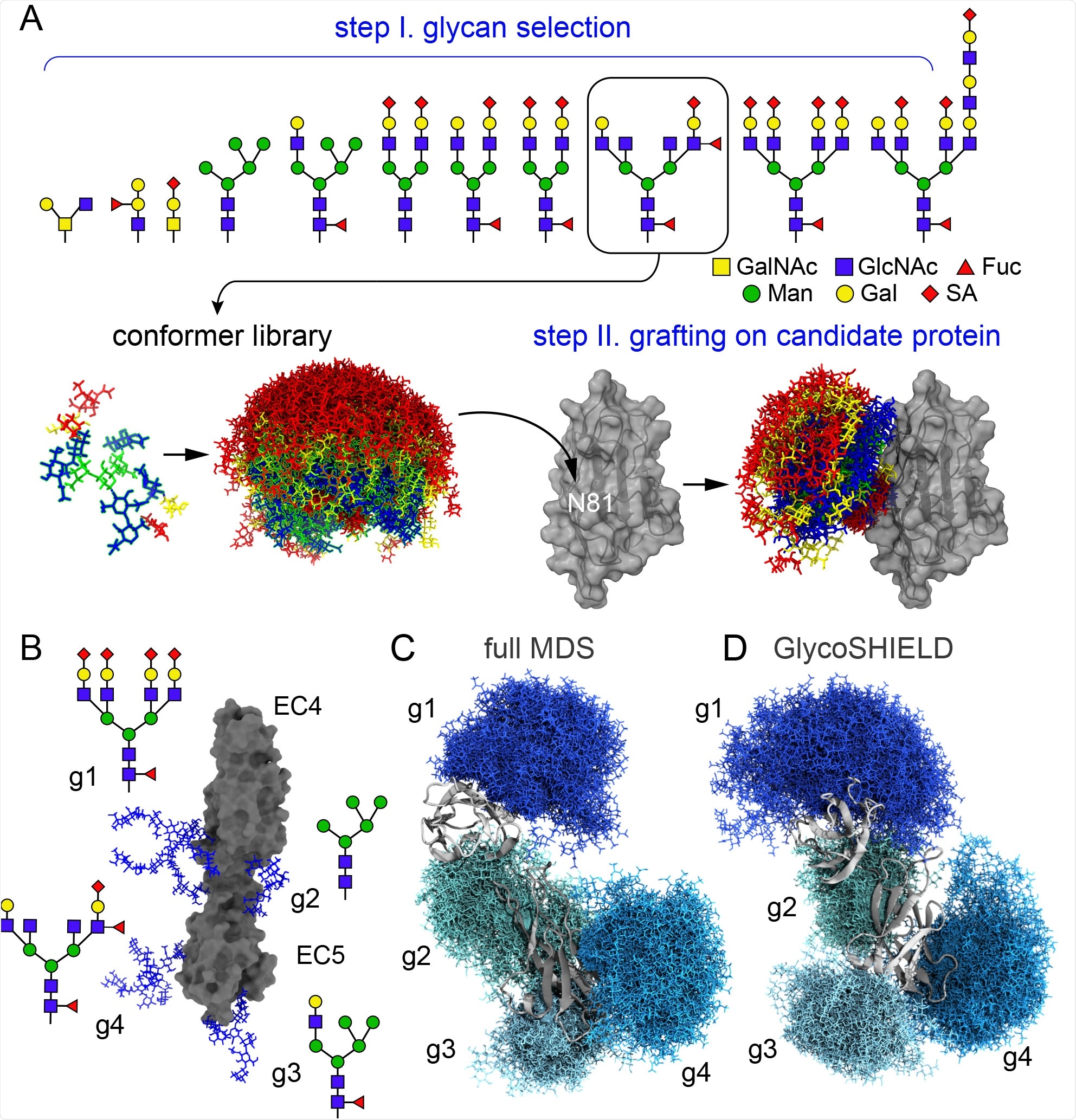
GlycoSHIELD generates realistic glycan shields A) Overview of the pipeline: user provides a 3D protein structure with defined glycosites where glycans from the library of conformers not clashing with the protein are grafted and exported for visualization and analysis (GalNac: N-Acetylgalactosamine, GlcNac: N-Acetylglucosamine, Fuc: fucose, Man: mannose, Gal: galactose, SA: sialic acid). B) Structure of N-cadherin EC4-EC5 model system with four distinct N-glycans as indicated at each glycosylation site (g1-g4). C) -D) Glycan conformers generated by full MDS (C) or with GlycoSHIELD (D) after alignment on EC4-EC5. Note the comparable morphology and span of the glycan shields obtained by the two approaches.
Water interacts with glycans when in physiological conditions, and the degree of interaction between glycans and the solvent can be estimated by the number of hydrogen bonds.
A similar number of hydrogen bonds were observed in the MDS and GlycoSHIELD simulations, indicating that the slight difference in protein shielding noted was likely due to imperfect glycan structure simulation in the latter, not having the necessary variety in conformational structure to account for the flexibility of the protein surface.
The internal motion of N-cadherin was essentially unchanged by either form of glycosylation, and the group concludes that GlycoSHIELD matches the expected estimates of glycan shielding and its influence on protein conformation from more sophisticated computational methods.
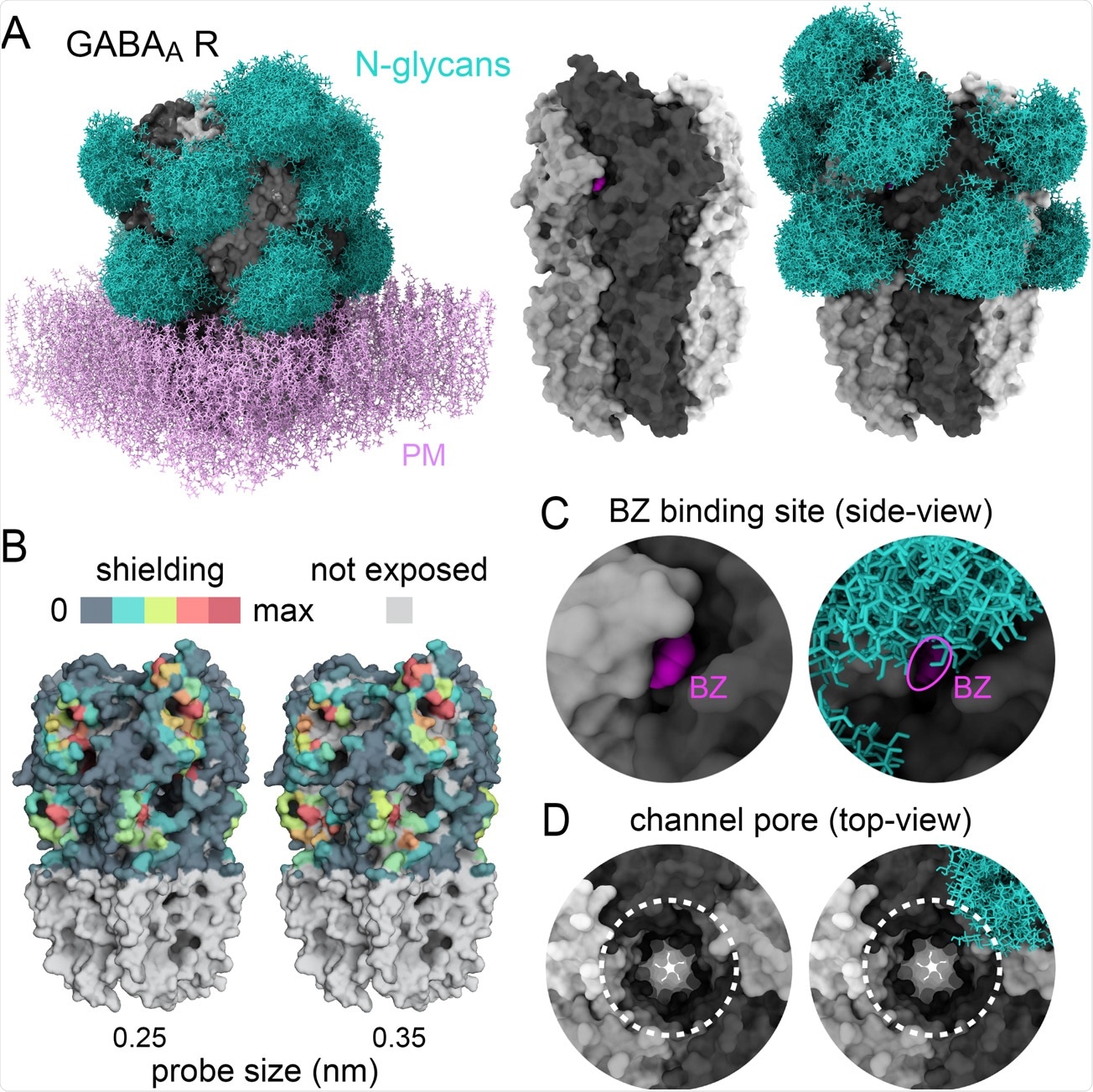
Glycans sculpt the surface of GABAA receptors A) Structure of a human homopentameric GABAA receptor (beta3 subunits, grey) shown with and without patch of plasma membrane (PM, added for visualisation only, pink) with and without reconstituted glycan shields (GlcNac2-Man5, blue in A and B). B) 3D-heatmaps of the reduction of solvent accessible surface area (SASA) by glycans for different probe size. C-D) Magnified views of the benzodiazepine binding site with bound ligand, BZ, shown in fuchsia (C) and the extracellular vestibule of the channel pore (dotted lines in D). In C and D, note the partial occlusion of the ligand binding site and the ending of the glycan shield at the opening of the channel pore, respectively.
SARS-CoV-2 spike protein glycosylation
The authors next applied GlycoSHIELD to the SARS-CoV-2 spike protein, which has been the focus of intense scrutiny since the start of the COVID-19 pandemic.
Glycosylation of the spike protein has been found by many other studies to play an important role in the way in which it interacts with neutralizing antibodies and receptors, including the target host receptor ACE2, and therefore accurate modeling is essential. The glycosylated spike protein obtained using GlycoSHIELD was again highly representative of the protein shield obtained using MDS techniques and captured how epitopes are masked.
One hundred sixty glycan conformers were utilized in this case, mapped on a best-fit basis to the glycosylation sites of the spike protein. Areas of least agreement between the two methods were around the loops of the receptor-binding domain, attributed to the short simulation time employed, as significant conformational changes can occur in this region. Thus more time is required to equilibrate. On stable protein structures, however, GylcanSHIELD produced highly realistic glycan shields.
Further, the group states that GlycanSHIELD can predict the structural impact of glycosylation on some proteins. For example, the SARS-CoV-2 spike protein stalk is highly flexible and stands more upright when glycosylated, as demonstrated by cryo-EM and computational methods. Having undergone glycosylation by GlycanSHIELD, the stalk was noted to be conformationally more rigid, suggesting that GlycanSHIELD can also be used to predict key structural features of proteins resulting from the addition of glycans.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.