By retrieving 329,942 severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) records from the GISAID database, researchers from a software company in Cambridge described genetic variability of SARS-CoV-2 in order to inform further studies. Their findings are currently available on the bioRxiv* preprint server while it undergoes peer review.
Over a year and a half into the pandemic, we are still not completely aware why exactly there is such high transmissibility of SARS-CoV-2, a causative agent of the coronavirus disease 2019 (COVID-19). Furthermore, the clinical presentation of an infection can vary greatly between individuals irrespective of several recognized risk factors.
Genetic recombination across host species boundaries is one of the key characteristics of SARS-CoV-2. As a result, the genome of this virus harbors signatures denoting multiple recombination events, presumably incorporating manifold species and wide geographic regions.
Hence, the analyses of single nucleotide polymorphisms are particularly beneficial in unveiling heavily mutated genomes and comprehending viral changing patterns. More specifically, relationship patterns between single nucleotide polymorphisms and their consequences can be forecasted from genotyping, transmission tracking and protein analysis.
Consequently, as common knowledge of SARS-CoV-2 signaling pathways, protein functioning, as well as the interaction of proteins with cells and other proteins, continues to rapidly accumulate due to its novelty, there is a pressing need to explore the SARS-CoV-2 changes.
This was recently pursued by researchers from Quantori software company in Cambridge (USA) and Mental Health Research Center in Moscow (Russia), as they claim that the key to understanding the global success of SARS-CoV-2 is hidden in its genome.
A deep dive into the GISAID database
This research group has analyzed 329,942 SARS-CoV-2 records obtained from the GISAID database, which is the world's most comprehensive publicly accessible database where both influenza and (since the start of pandemic) SARS-CoV-2 sequence data are stored.
The scientist has addressed the quality of the records uploaded to the aforementioned database, gender distribution, gene conservation, single nucleotide polymorphisms, clusters, insertions and deletions, and a correlation coefficient matrix.
Genomic coordinates were acquired from the University of California Santa Cruz (UCSC) Genome Browser, a web-based open-source graphical viewer to display genome sequences and their annotations. Furthermore, sequence alignments were performed for every gene separately.
Finally, the completeness of the GISAID database was also addressed by the researchers, as many fields were filled incorrectly or left blank. Metadata mining analysis has resulted in a hypothesis on gender inequality in medical care in certain countries.
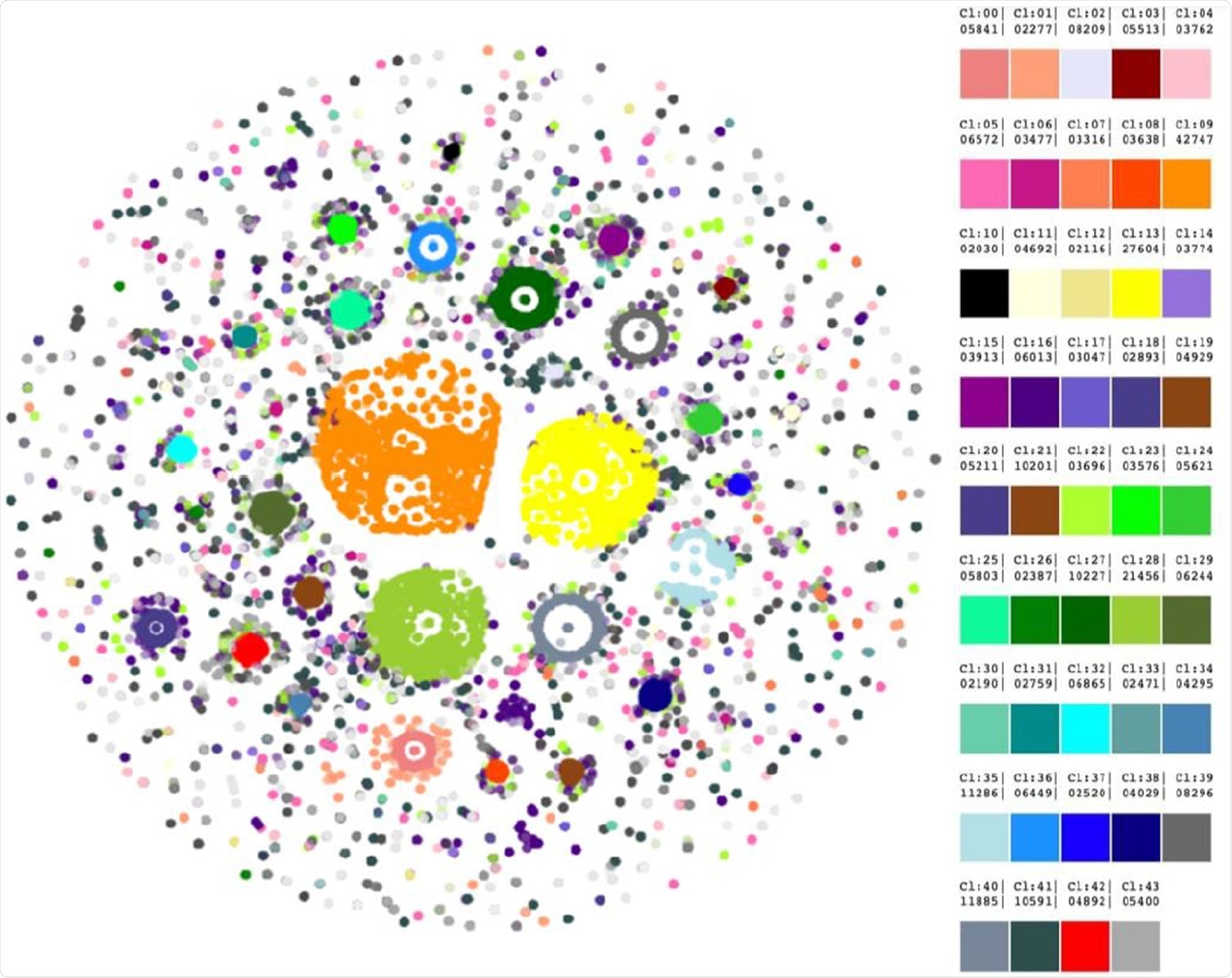
Forty-three clusters were revealed by HDBScan. Legend on the right contains cluster numbers and color schemes.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
A potential emergence of new variants
This study has shown that mutations arising with high frequency (i.e., more than 0.3%) are not abundant and account for 155 changes when all genes are concerned. Moreover, many mutations harbored concomitant changes that could alter the consequences for the virus or a human host.
An important caveat is that most data are from the United Kingdom (UK), which results in an overall data bias towards the UK statistics. However, it was still visible that two clusters were formed by mutations found in samples uploaded mainly by Australia and Denmark, indicating a potential emergence of "Australian" or "Danish" SARS-CoV-2 variants.
Furthermore, conservation analysis pinpointed ORF6 and E genes as possible treatment or vaccine targets due to their high conservation. There is also a possibility of the existence of a subtype of a B.1.1.7 variant, previously known as the UK variant.
The need for further research
Despite the daily accumulation of a vast amount of knowledge, the exact consequences of many viral mutations are thus far unknown. Still, an abundance of co-occurring mutations prompt us to pursue additional research on their meaning and potential of creating new variants.
"Taken together, our results describe the genetic variability of SARS-CoV-2 and may be used for further research in different scientific areas", say study authors in this bioRxiv paper. "Our results indicate areas of the SARS-CoV-2 genome that researchers can focus on for further structural and functional analysis", they conclude.
In any case, such unprecedented data growth and surge in expert analyses provide a myriad of opportunities to develop specific science-guided policies that will aid in the design and implementation of stringent epidemiological practices for preventing future outbreaks.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Zelenova, M., Ivanova, A., Semyonov, S. & Gankin, Y. (2021). Analysis of 329,942 SARS-CoV-2 records retrieved from GISAID database. bioRxiv. https://doi.org/10.1101/2021.08.04.454929, https://www.biorxiv.org/content/10.1101/2021.08.04.454929v1
- Peer reviewed and published scientific report.
Zelenova, Maria, Anna Ivanova, Semyon Semyonov, and Yuriy Gankin. 2021. “Analysis of 329,942 SARS-CoV-2 Records Retrieved from GISAID Database.” Computers in Biology and Medicine 139 (December): 104981. https://doi.org/10.1016/j.compbiomed.2021.104981. https://www.sciencedirect.com/science/article/pii/S0010482521007757.