The rapid outbreak of the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) has caused the current coronavirus disease 2019 (COVID-19) pandemic. Scientists around the world have been working extremely hard to contain the pandemic by developing effective vaccines and therapeutics, formulating non-pharmaceutical measures, and screening for the emergence of new SARS-CoV-2 variants.
Linear epitopes are categorized as T-cell epitopes and B-cell epitopes. T-cell epitopes bind to the major histocompatibility complex (MHC) molecules found on the cell surface, while B-cells bind to secreted and cell-bound immunoglobulins. Typically, in addition to containing B-cell or T-cell epitopes, epitope-based vaccines contain multiple distinct epitopes to enhance their effectiveness, especially during the evolution of the virus.
The nucleocapsid (N) protein is one of the main components of SARS-CoV-2 and encapsulates the negative-stranded ribonucleic acid (RNA). The SARS-CoV-2 N protein is also involved with viral functions, including messenger RNA (mRNA) transcription, replication, and immune regulation. The SARS-CoV-2 N protein contains two functionally distinct and conserved domains, including the N-terminal RNA-binding domain (NTD) and the C-terminal domain (CTD).
The SARS-CoV-2 spike (S) protein also plays a vital role in virus binding, fusion, and entry into the host cell. The S protein is a homotrimeric class I fusion protein that contains the S1 and S2 domains.
About the study
In a recent bioRxiv* study, researchers identify and investigate the presence and evolution of the SARS-CoV-2 epitopes, particularly the N and S glycoprotein proteins, in order to advance the current understanding of the adaptive immune response to SARS-CoV-2. In this study, the researchers analyzed eleven different SARS-CoV-2 proteins that resulted in twenty-eight thousand unique protein sequences.
A total of 112 laboratory-confirmed B-cell epitopes were identified, three of which were found on SARS-CoV-2, while others were found in SARS-CoV. Additionally, 279 lab-confirmed T-cell epitopes were identified, among which 221 were found on SARS-CoV-2, while the remaining epitopes belonged to SARS-CoV.
Based on a common set of epitopes between SARS-CoV-2 and other viruses within the Coronaviridae family, the researchers performed hierarchical clustering of the epitopes to detect epitopes that overlap the same protein region. Prediction of previously unknown epitopes and the identification of their location on specific proteins can determine which epitopes are evolving rapidly. These efforts can also identify which epitopes are more stable as potential candidates for vaccine targets.
Study findings
Complete sequence homology was shared only between SARS-COV-2 and SARS epitopes. While performing an exact sequence match, no epitopes from other Coronaviridae were present in SARS-COV-2 genomes.
Although no exact match of epitopes was obtained with Coronaviridae other than SARS-CoV and SARS-CoV-2, B-cell epitopes contained parent epitopes that were similar to other organisms, such as the murine hepatitis virus strain JHM, Feline infectious peritonitis virus (strain KU-2), infectious bronchitis virus, and porcine epidemic diarrhea virus.
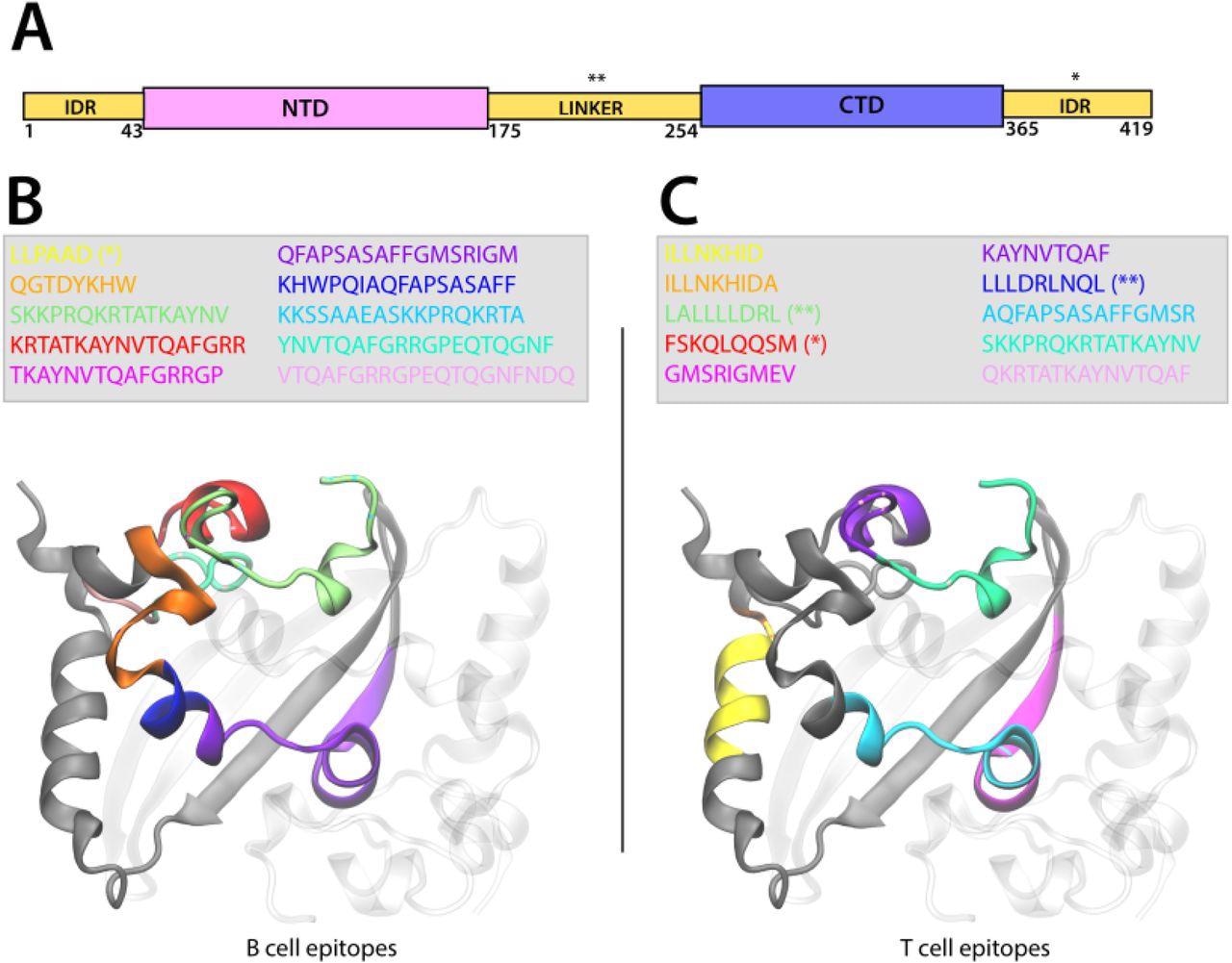
Representation of the localization of the B-Cell and T-Cell epitopes on the CTD domain of the Nucleoprotein. A. Scheme of SARS-CoV-2 N domains illustrating the N-term intrinsically disordered region (IDR) followed by the N-terminal domain (NTD), the IDR linker, the C-terminal domain (CTD), and the C-term IDR. B-C. The N CTD dimer is represented in New Cartoon format (one monomer is gray-colored and the other is transparent) and the sequence of the B-Cell (B) and T-Cell (C) epitopes is colored according to the legend represented in the figure. The epitope sequence is represented in the legend. The epitopes located in the linker domain are indicated by (**) and those in the C-term IDR by (*). For great clarity, we represented the epitopes in only one monomer.
Additionally, novel T-cell epitopes were also found to have parent epitopes belonging to organisms other than SARS and SARS-COV-2, including the feline infectious peritonitis virus strain KU-2 and murine hepatitis virus. Identification of these epitopes could play an essential role in developing novel therapeutics.
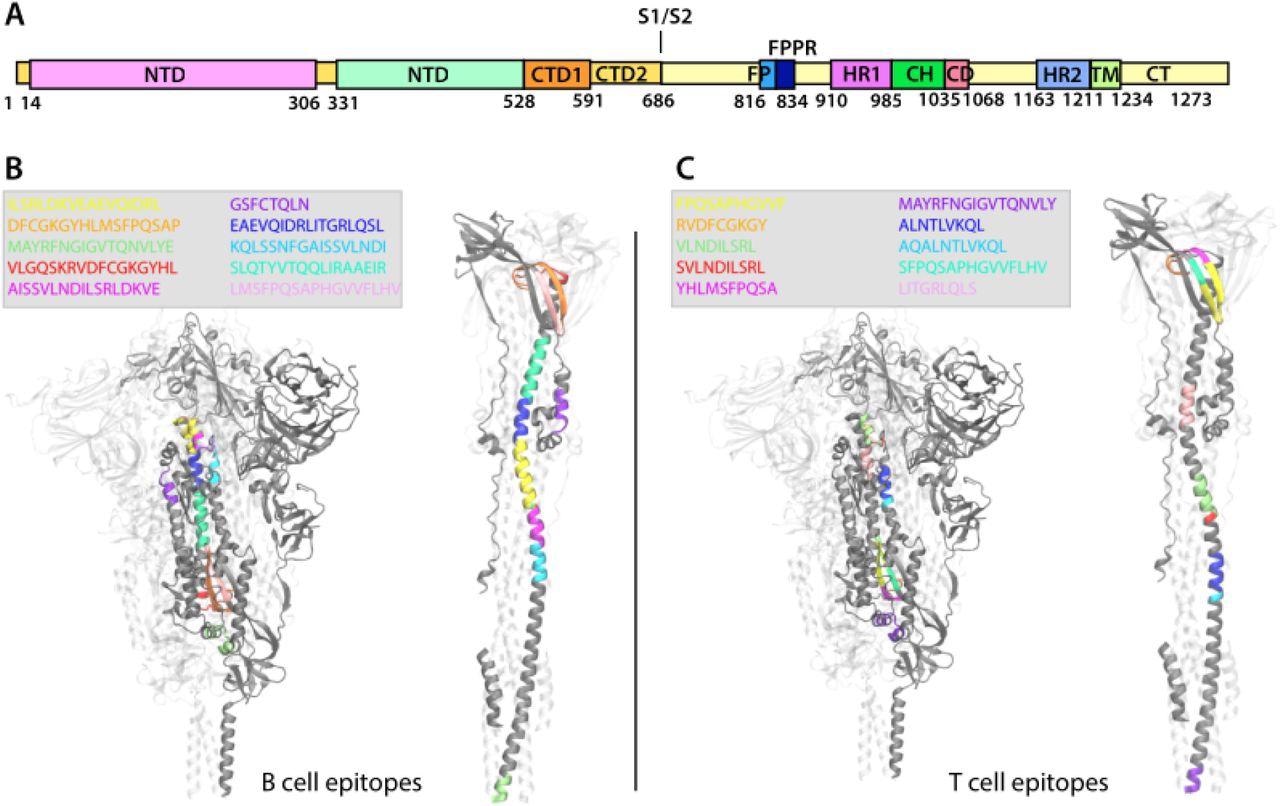
Representation of the localization of the B-Cell and T-Cell epitopes on the SARS-CoV-2 Spike glycoprotein in the prefusion and postfusion conformations. A. Scheme of SARS-CoV-2 S1 and S2 units of the S protein and of their domains. B-C. The S protein trimer is represented in New Cartoon format (one monomer is gray-colored the other two are transparent) and is shown in the prefusion conformation on the left side of the panels and in the postfusion conformation on the right side of the panels. The sequence of the B-Cell (A) and T-Cell (B) epitopes is shown in the figure legend and is colored accordingly in the S protein structure.
Among the top ten epitopes for each epitope type and protein identified, the most frequent epitopes have parent epitopes belonging to SARS-CoV-2 and SARS-CoV, except for one T-cell Spike glycoprotein epitope. An extensive set of epitopes have been detected that could be used for effective vaccine development.
The scientists also measured the evolution rate of epitopes based on sequence-based clustering plots, which could help select the most suitable epitopes for vaccine development. Additionally, evaluation of mutation density regions and immunodominance regions helped predict epitopes that may undergo the least changes in their amino acids.
In the three-dimensional representation of the N and the S proteins, these epitopes were found to be mostly localized in the CTD of the N protein, whereas their location in the S protein was primarily confined to the S2 subunit.
Conclusions
Identifying the position of the most commonly conserved epitopes and glycan sites is critical for developing wind-spectrum vaccines and therapeutics.
In the future, the authors of the current study will analyze the proximity of epitopes to more mutation-prone regions, which would help narrow down more potential epitopes for vaccine development. The researchers also hope to design quantitative metrics for ranking epitope targets suitable for vaccine design and development.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Agarwal, A., Beck, K. L., Capponi, S., et al. (2022) Predicting Epitope Candidates for SARS-CoV-2. bioRxiv. doi:10.1101/2022.02.09.479786. https://www.biorxiv.org/content/10.1101/2022.02.09.479786v1.
- Peer reviewed and published scientific report.
Agarwal, Akshay, Kristen L. Beck, Sara Capponi, Mark Kunitomi, Gowri Nayar, Edward Seabolt, Gandhar Mahadeshwar, Simone Bianco, Vandana Mukherjee, and James H. Kaufman. 2022. “Predicting Epitope Candidates for SARS-CoV-2.” Viruses 14 (8): 1837. https://doi.org/10.3390/v14081837. https://www.mdpi.com/1999-4915/14/8/1837.