Testing for SARS-CoV-2 is important for monitoring and controlling COVID outbreaks since it allows healthcare authorities to manage SARS-CoV-2-infected persons appropriately based on their test-trace results. In addition, viruses released into the sewage networks by the general population can be detected and quantified at specific periods.
The presence of SARS-CoV-2 particles in the feces of people infected with the virus correlates with the amount detected in COVID-19 community testing. Therefore, sewage surveillance can monitor the extent of SARS-CoV-2 transmission in the community and has the potential to detect SARS-CoV-2 outbreaks at an early stage.
About the study
In the present study, researchers described tracking of SARS-CoV-2 RNA levels by the WBE approach based on the near-real-time dataset of a nationwide program conducted by Scottish Water and the Scottish environmental protection agency (SEPA) between May 2020 and February 2022.
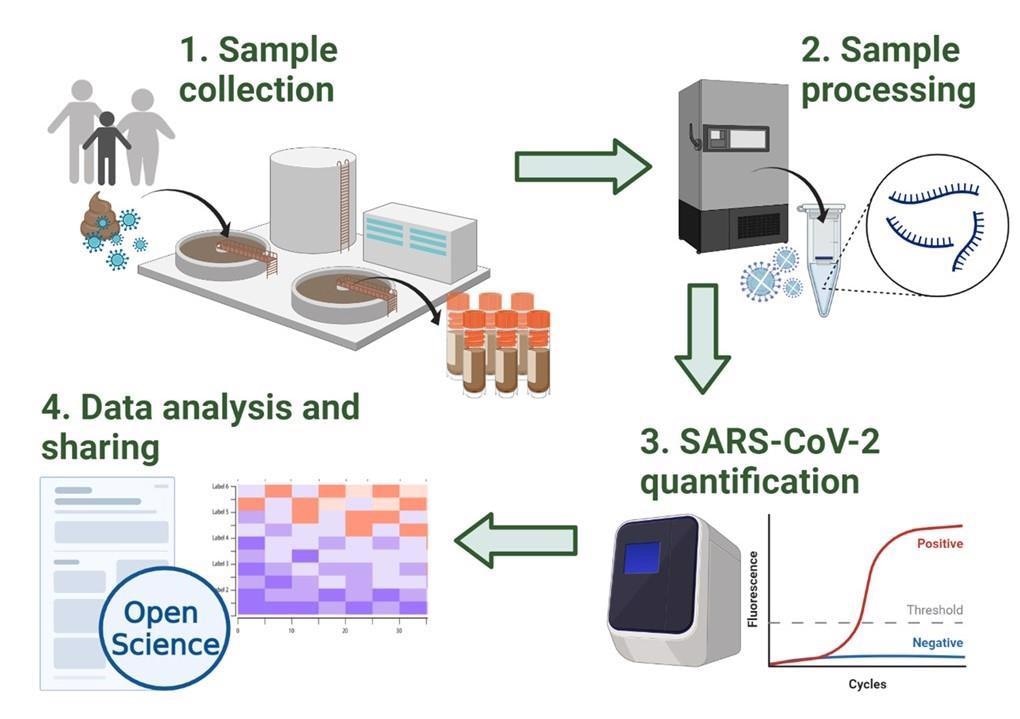
Overview of the methodology workflow of the Scottish SARS-CoV-2 monitoring in wastewater program. Incoming wastewater samples are collected by Scottish Water (1) and transferred to SEPA’s laboratories where samples are stored and processed (2). SARS-CoV-2 viral levels are then quantified using RT-qPCR (3). Finally, the data obtained is shared on SEPA’s public dashboard and with local authorities such as the Scottish Government. Additionally, the detailed methodology and datasets are shared in open online platforms and repositories such as protocols.io, Zenodo and GitHub 19–21,26,27. This figure was created with BioRender.com.
Wastewater samples were collected by the Scottish Water (SW) team of operators from 122 sites in 14 national health service (NHS) Scotland health board regions and transferred to SEPA laboratories for storage and further processing. SARS-CoV-2 RNA was extracted from the samples and quantified using quantitative reverse transcription-polymerase chain reaction (RT-qPCR) assays of the SARS-CoV-2 nucleocapsid 1 (N1) gene.
Samples were collected once or twice per week, up to four times per week, using a 24-hour-autosampler or a grab sample from maintenance holes. Before processing, the wastewater samples were spiked with a known target of non-target RNA of porcine reproductive and respiratory syndrome (PRRS) virus as a quality control measure. The cycle threshold (Ct) values and the gene copies per liter (gc/L) values were calculated for each sample.
Data obtained were shared on public dashboards of SEPA and with the government of Scotland. In addition, the datasets were shared in open online platforms and repositories such as GitHub, protocols.io, and Zenodo. Data were normalized for the final analysis, based on information such as the site population, incoming wastewater flow values, and ammonia content, and expressed as million gene copies per person per day [Mgc/pD]. Statistical analysis was performed using the biomathematics and statistics Scotland (BioSS) and the data were presented in the Scottish government reports, published weekly.
Based on the number of N1 gene replicates per liter, the wastewater samples were classified as negative, weakly positive, positive detected, not quantifiable (positive DNQ), or positive. If ≥2 replicates didn’t produce Ct, the sample was reported to be negative. If two replicates from three replicates or one replicate from two replicates demonstrated positive signals, but the signals were lower than the limit of detection (LoD), the sample was reported to be weakly positive. If the mean values of replicates were between the LoD (1,316 gc/L) and the limit of quantification (LoQ, 11,386 gc/L) but could not be the quantity was not statistically significant, the wastewater sample was reported to be positive DNQ. If the mean values of all replicates exceeded the LoQ value, the sample was reported to be positive.
Results
In the analysis, 9.3%, 7.3%, 14%, and 63.4% of the samples were reported as negative, weakly positive, positive DNQ, and positive, respectively. However, six percent of the study samples could not be categorized. Data normalization showed that data obtained by the WBE approach correlated well with data published by local health authorities on the SARS-CoV-2 pandemic waves.
![Map of Scotland showing sites from which wastewater samples were collected for SARS-CoV-2 analysis. The colored circles show normalized SARS-CoV-2 virus levels on the last week of July 2021 in Million gene copies per person per day ([Mgc/pD]; truncated to 90). The grey circles represent sites without measurement on that week. See Methods – Data visualization for further details.](https://www.news-medical.net/images/news/ImageForNews_716756_1655262601818580.jpg)
Map of Scotland showing sites from which wastewater samples were collected for SARS-CoV-2 analysis. The colored circles show normalized SARS-CoV-2 virus levels on the last week of July 2021 in Million gene copies per person per day ([Mgc/pD]; truncated to 90). The grey circles represent sites without measurement on that week. See Methods – Data visualization for further details.
A nationwide increase in COVID-19 cases was reported in the period between December 2020 and February 2021, which was followed by fewer COVID-19 cases between February 2021 and mid-June 2021, after which COVID-19 cases began to increase again. Similar COVID-19 trends were also observed after evaluating the SARS-CoV-2 RNA levels in wastewaters by the WBE approach.
Likewise, COVID-19 cases peaked in September 2021, following which the cases dropped and again bounced back with another peak between December end 2021 and February 2022, which correlated with a directly proportional increase and decrease in SARS-CoV-2 RNA levels in the wastewaters in Scotland, United Kingdom (UK). Moreover, in the quality control experiments, PRRS-causing viral pathogen was successfully detected indicating that the study’s methodology was accurate and also that a negative test was most likely due to the SARS-CoV-2 N1 gene absence in the wastewater samples and not due to failures in methodology.
Overall, the findings showed that mathematical modeling of wastewater data could be used to estimate daily counts of COVID-19 cases and the nationwide prevalence of SARS-CoV-2 infections. The WBE approach can be a helpful aid in detecting predicting surges of SARS-CoV-2 infections or similar viral infections in the future.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
SARS-CoV-2 RNA levels in Scotland’s wastewater. Livia C. T. Scorza, Graeme J. Cameron, Roisin Murray-Williams, David Findlay, Julie Bolland, Brindusa Cerghizan, Kirsty Campbell, David Thomson, Alexander Corbishley, David Gally, Stephen Fitzgerald, Alison Low, Sean McAteer, Adrian M. I. Roberts, Zhou Fang, Claus-Dieter Mayer, Anastasia Frantsuzova, Sumy V. Baby, Tomasz Zieliński, Andrew J. Millar. medRxiv preprint 2022, DOI: https://doi.org/10.1101/2022.06.08.22276093, https://www.medrxiv.org/content/10.1101/2022.06.08.22276093v1
- Peer reviewed and published scientific report.
Scorza, Livia C. T., Graeme J. Cameron, Roisin Murray-Williams, David Findlay, Julie Bolland, Brindusa Cerghizan, Kirsty Campbell, et al. 2022. “SARS-CoV-2 RNA Levels in Scotland’s Wastewater.” Scientific Data 9 (1): 713. https://doi.org/10.1038/s41597-022-01788-3. https://www.nature.com/articles/s41597-022-01788-3.