The rapid spread of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) led to the coronavirus disease 2019 (COVID-19) pandemic, which has, to date, claimed more than 6.4 million lives globally.
The emergence of new SARS-CoV-2 variants due to the development of genomic mutations has prolonged the pandemic. The Delta and Omicron variants of concern (VOCs), for example, can escape immune protection elicited through both COVID-19 vaccination and natural infection.

Study: Monovalent and trivalent VSV-based COVID-19 vaccines elicit potent neutralizing antibodies and immunodominant CD8+ T cells against diverse SARS-CoV-2 variants. Image Credit: Alberto Andrei Rosu / Shutterstock.com

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
All currently available COVID-19 vaccines are based on the spike protein of the original SARS-CoV-2 strain. Due to the reduced efficacy of COVID-19 vaccines against current SARS-CoV-2 variants, there is an urgent need for efficient, safe, and cost-effective vaccines capable of protecting the human population from new variants.
The vesicular stomatitis virus (VSV) belongs to the family Rhabdoviridae, which has been effectively used to express antigens on its surface. VSV is a safe vaccine platform that is either used alone or in combination with or replacing the VSV envelope glycoprotein to develop an immunogenic vaccine vector. The immunogenic vector triggers an immune response in the mucosa.
As compared to other viral vector platforms, such as adenoviruses, VSV rarely infects humans; therefore, pre-existing seropositivity is low in the human population. This is advantageous, as it substantially reduces the effect of anti-vector immunity, which can cause hindrance to repeated booster vaccination.
Another advantage of the VSV platform is that it can maintain attenuated replication, which enables antigenic presentation for a long duration and minimizes vaccine dose requirements. Additionally, this vaccine approach reduces the cost of vaccine production as compared to other vaccine vectors.
Ervebo is a recombinant VSV (rVSV) vaccine that was developed against the Ebola virus and is the first of its kind to receive approval from the U.S. Food and Drug Administration (FDA). During the COVID-19 pandemic, many scientists looked to the rVSV vaccine platform to develop potential COVID-19 vaccines.
Previous studies have reported the development of SARS-CoV-2-specific rVSV vaccines based on the ancestral SARS-CoV-2 spike protein that elicited spike-specific T-cell responses and neutralizing antibodies (nAbs). Moreover, in vivo experiments revealed that this vaccine can effectively protect animals from severe infection caused by the original SARS-CoV-2 strain as well as elicit rapid viral clearance.
About the study
A new study published on the bioRxiv* preprint server discusses the development of rVSV COVID-19 vaccines and determined their efficacy against the original SARS-CoV-2 strain, as well as the Beta, Delta, and Omicron variants.
The rVSV-SARS-CoV-2 variant vaccines were constructed against the Beta and Delta variants. To this end, the mutations of these variants were incorporated into the human codon-optimized full-length spike (SF) protein of SARS-CoV-2.
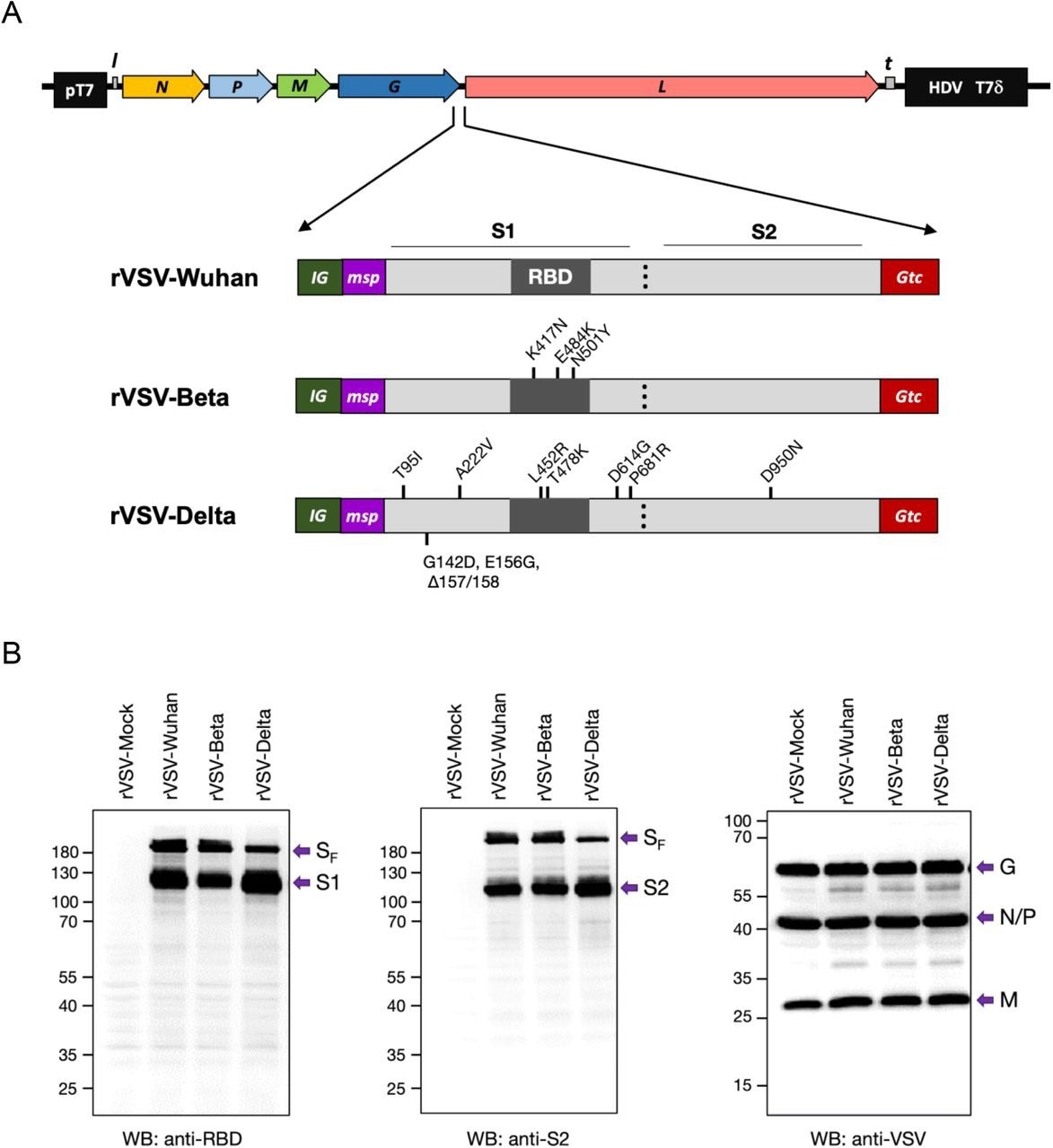 Construction of recombinant rVSV with SF genes of SARS-CoV-2 Wuhan, Beta, and Delta. (A) Constructs of VSV-SARS-CoV-2 Wuhan (rVSV-Wuhan), Beta (rVSV-Beta), and Delta (rVSV-Delta). Codon-optimized full-length Spike protein gene (SF) with VSV intergenic sequences (IG), 21 amino acid honeybee melittin signal peptide (msp), and 49 amino acid VSV G protein transmembrane domain and cytoplasmic tail (Gtc) were inserted into the G and L gene junction of rVSVInd. (B) Expression of SARS-CoV-2 Spike proteins in BHK-21 cells at 6 hr post-infection with rVSV-Mock (rVSV), rVSV-Wuhan, rVSV-Beta, and rVSV-Delta. pT7: Bacteriophage T7 promoter for DNA dependent RNA polymerase. N: VSV Nucleocapsid protein gene. P: VSV Phosphoprotein gene. M: VSV Matrix protein gene. G: VSV Glycoprotein gene. L: VSV Large protein, RNA dependent RNA polymerase gene. l: Leader region in the 3’ s-end of the VSV genome. t: Trailer region in the 5’-end of the VSV genome. HDV: Hepatitis delta virus ribozyme encoding sequences. T7δ: Bacteriophage T7 transcriptional terminator sequences. nt: nucleotides. aa: amino acids.
Construction of recombinant rVSV with SF genes of SARS-CoV-2 Wuhan, Beta, and Delta. (A) Constructs of VSV-SARS-CoV-2 Wuhan (rVSV-Wuhan), Beta (rVSV-Beta), and Delta (rVSV-Delta). Codon-optimized full-length Spike protein gene (SF) with VSV intergenic sequences (IG), 21 amino acid honeybee melittin signal peptide (msp), and 49 amino acid VSV G protein transmembrane domain and cytoplasmic tail (Gtc) were inserted into the G and L gene junction of rVSVInd. (B) Expression of SARS-CoV-2 Spike proteins in BHK-21 cells at 6 hr post-infection with rVSV-Mock (rVSV), rVSV-Wuhan, rVSV-Beta, and rVSV-Delta. pT7: Bacteriophage T7 promoter for DNA dependent RNA polymerase. N: VSV Nucleocapsid protein gene. P: VSV Phosphoprotein gene. M: VSV Matrix protein gene. G: VSV Glycoprotein gene. L: VSV Large protein, RNA dependent RNA polymerase gene. l: Leader region in the 3’ s-end of the VSV genome. t: Trailer region in the 5’-end of the VSV genome. HDV: Hepatitis delta virus ribozyme encoding sequences. T7δ: Bacteriophage T7 transcriptional terminator sequences. nt: nucleotides. aa: amino acids.
The scientists also introduced honeybee melittin signal peptide (msp) to replace the SF amino terminus and VSV G protein transmembrane domain. Similarly, the cytoplasmic tail (Gtc) replaced the SF carboxy terminus.
The newly developed rVSV-Beta vaccine contained several mutations, including E484K, K417N, and N501Y in the receptor-binding domain (RBD) of codon-optimized SF. Similarly, the rVSV-Delta vaccine contained two amino acid changes at F157 and R158, as well as nine amino acid changes, including A222V, D614G, E156G, P681R, G142D, T95I, L452R, D950N, and T478K in the codon-optimized SF. The scientists also synthetically generated SF genes with mutations and incorporated them in between the glycoprotein (G) and polymerase (L) genes of VSV.
Six female mice received prime immunization through the intramuscular route using the rVSV-Wuhan vaccine, -Beta vaccine, -Delta vaccine, or trivalent formulation that contained an equal ratio of rVSV-Wuhan, rVSV-Beta, and rVSV-Delta vaccines. Three weeks after prime immunization, immunized mice were subjected to booster vaccination using monovalent or trivalent vaccines. Subsequently, blood samples were collected two weeks post booster vaccination and studied.
Study findings
Both monovalent rVSV-Wuhan and rVSV-Delta vaccines, as well as the trivalent vaccine, elicited effective nAbs and spike-specific CD8+ T-cell responses. These vaccines could effectively inhibit the original SARS-CoV-2 strain, as well as Beta, Delta, and Omicron variants.
Furthermore, the monovalent rVSV-Wuhan vaccine booster elicited immune responses that restricted both the original and Omicron strains. Booster vaccination with rVSV-Beta was less effective as compared to the rVSV-Wuhan vaccine.
A heterologous booster vaccination scheme that used the rVSV-Wuhan prime vaccine with a rVSV-Delta booster exhibited a significantly higher production of nAbs that were effective against the Delta and Omicron variants. Heterologous vaccination using the prime rVSV-Wuhan vaccine with a rVSV-trivalent booster dose also induced the production of nAbs and CD8+ T-cell responses that were effective against all tested SARS-CoV-2 variants.
![Immunization with monovalent and trivalent vaccines elicit a broad neutralizing antibody response. (A) Female BALB/c mice were prime immunized with 5×108 PFU intramuscularly (i.m) of either rVSV-monovalent (rVSV-Mock [M], rVSV-Wuhan [W], rVSV-Beta [B], rVSV-Delta [D]) or trivalent (rVSV-Wuhan-Beta-Delta [T]) vaccine (day 0). On day 21 mice were administered a 5×108 PFU homologous or heterologous boost i.m. Two weeks post-boost blood and spleen were collected for analysis of the immune response. (B) SARS-CoV-2 neutralization (SARS-CoV-2Wuhan_USAWA1, SARS-CoV-2Beta, SARS-CoV-2Delta, SARS-CoV-2Omicron) by immune sera two weeks post-boost was determined using replication-competent SARS-CoV-2 isolates in microneutralization assays. Graphed data are presented as arithmetic mean of log-transformed 50% neutralization titer (NT50). The horizontal dotted lines indicate the limit of detection. Each symbol denotes an individual animal;](https://www.news-medical.net/images/news/ImageForNews_720286_16586189040767294.jpg)
Immunization with monovalent and trivalent vaccines elicit a broad neutralizing antibody response. (A) Female BALB/c mice were prime immunized with 5×108 PFU intramuscularly (i.m) of either rVSV-monovalent (rVSV-Mock [M], rVSV-Wuhan [W], rVSV-Beta [B], rVSV-Delta [D]) or trivalent (rVSV-Wuhan-Beta-Delta [T]) vaccine (day 0). On day 21 mice were administered a 5×108 PFU homologous or heterologous boost i.m. Two weeks post-boost blood and spleen were collected for analysis of the immune response. (B) SARS-CoV-2 neutralization (SARS-CoV-2Wuhan_USAWA1, SARS-CoV-2Beta, SARS-CoV-2Delta, SARS-CoV-2Omicron) by immune sera two weeks post-boost was determined using replication-competent SARS-CoV-2 isolates in microneutralization assays. Graphed data are presented as arithmetic mean of log-transformed 50% neutralization titer (NT50). The horizontal dotted lines indicate the limit of detection. Each symbol denotes an individual animal; n=6; 2 independent experiments. Statistical significance was determined by one-way ANOVA with Bonferroni’s correction for multiple comparisons (*, p < 0.05; **, p<0.01; ***, p< 0.001; **** p<0.0001). Geometric mean NT50 (GMT) and the fold increases in GMT NT50 versus W/W (rVSV-Wuhan/Wuhan) are shown for each vaccine group.
Conclusions
In the current study, researchers report the development of replication-competent rVSV vaccines composed of SARS-CoV-2 variant spike proteins. Herein, they highlight the importance of rVSV variant spike vaccines as booster vaccination doses that could be effective against diverse SARS-CoV-2 variants.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Parham, A. K., Kim, G. N., Saeedian, N., et al. (2022) Monovalent and trivalent VSV-based COVID-19 vaccines elicit potent neutralizing antibodies and immunodominant CD8+ T cells against diverse SARS-CoV-2 variants. bioRxiv. doi:10.1101/2022.07.19.500626
- Peer reviewed and published scientific report.
Parham, Kate A., Gyoung Nyoun Kim, Connor G. Richer, Marina Ninkov, Kunyu Wu, Nasrin Saeedian, Yue Li, et al. 2023. “Monovalent and Trivalent VSV-Based COVID-19 Vaccines Elicit Neutralizing Antibodies and CD8+ T Cells against SARS-CoV-2 Variants.” IScience 26 (4): 106292. https://doi.org/10.1016/j.isci.2023.106292. https://www.sciencedirect.com/science/article/pii/S2589004223003693.