The coronavirus disease 2019 (COVID-19) pandemic has caused a significant perturbation of public health. Conventional biomolecular tests have been pivotal in tracking severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). MS-based metabolomics represents the latest innovative technologies to identify circulating metabolites as COVID-19 biomarkers. In the present study, researchers reviewed COVID-19-related metabolome for potential biomarkers.
 Study: Diagnostic, Prognostic and Mechanistic Biomarkers of COVID-19 Identified by Mass Spectrometric Metabolomics. Image Credit: Intothelight Photography / Shutterstock
Study: Diagnostic, Prognostic and Mechanistic Biomarkers of COVID-19 Identified by Mass Spectrometric Metabolomics. Image Credit: Intothelight Photography / Shutterstock
Prognostic COVID-19 biomarkers
Metabolomics has been leveraged in biomarker discovery to identify metabolites correlating with diseases. Multiple studies have evaluated COVID-19 metabolome by applying different criteria such as pathologic severity, outcome(s), risk factors for severe illness, treatments, and longitudinal analysis of infected individuals. These heterogeneous criteria may compromise comparisons between studies.
However, studies have adopted a robust clinical strategy to strengthen metabolomic analyses. Clinical standardization can be achieved with strict inclusion criteria and broad clinical characteristics. COVID-19 severity requires a more accurate distinction between mild, moderate, and severe illness. In addition, it is essential to assess the reproducibility of measurements and evaluate them in validation cohorts to confirm the identification of biomarkers.
In targeted and non-targeted metabolomics, chromatographic methods are coupled to mass spectrometers, with typical combinations including liquid chromatography (LC)-MS, gas chromatography (GC)-MS, and ultra-performance LC-MS (UPLC-MS). Untargeted metabolomics has boosted the discovery of potential COVID-19 biomarkers.
This approach facilitates the comprehensive detection of metabolites requiring subsequent validation by a targeted method. Various mass analyzers have been used to identify COVID-19 metabolic biomarkers, such as phenols, mannose compounds, fructose, purine nucleosides, free fatty acids, and amino acids, among others. Machine-learning algorithms have recently facilitated the detection of unique biomarkers.
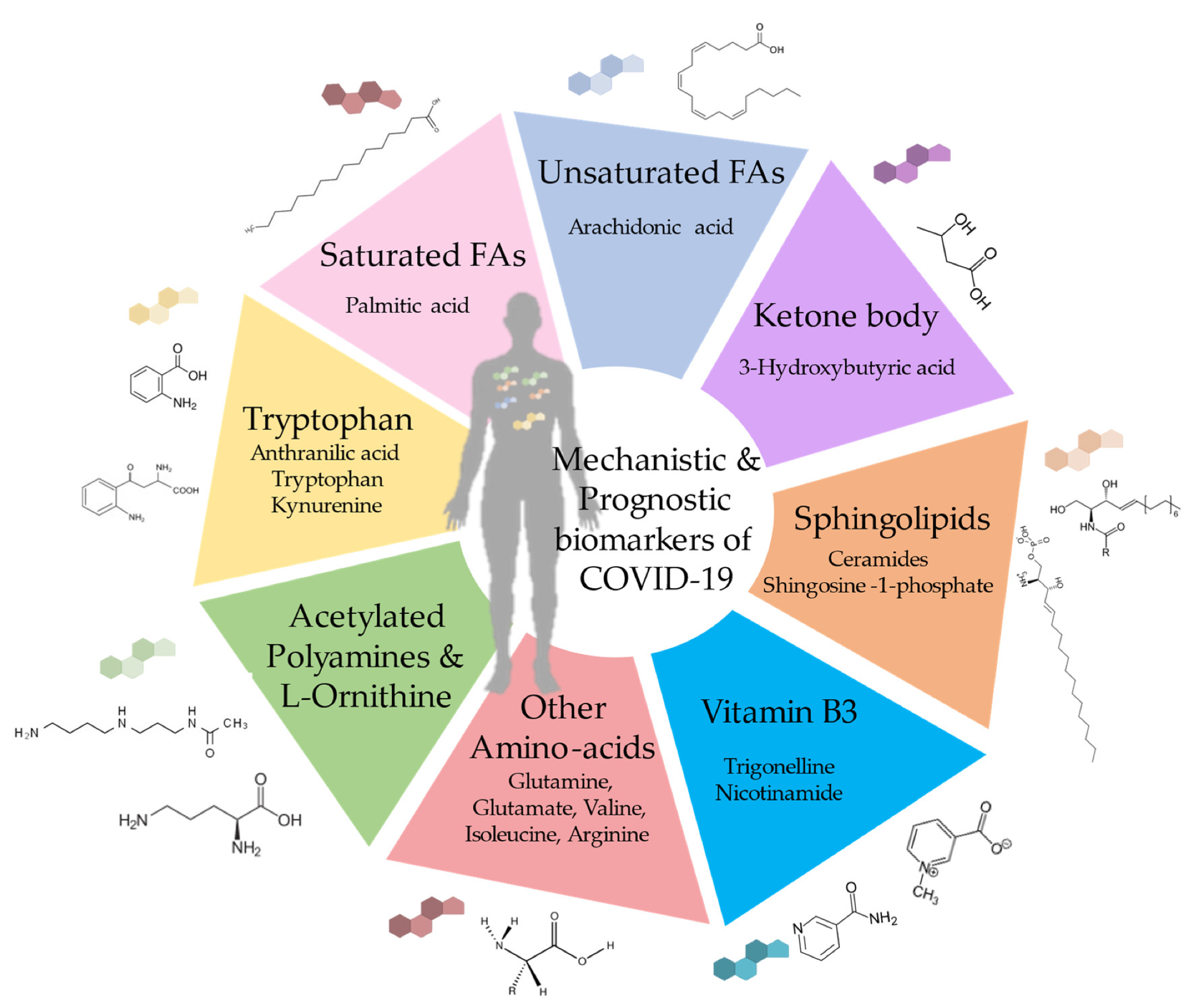 Mechanistic, diagnostic and prognostic biomarkers of COVID-19 disease.
Mechanistic, diagnostic and prognostic biomarkers of COVID-19 disease.
Diagnostic and prognostic features of COVID-19 metabolome
A literature search was performed for studies applying MS-based metabolomics in COVID-19. Eligible studies recruited SARS-CoV-2-infected individuals with a positive reverse-transcription polymerase chain reaction (RT-PCR) test and controls without COVID-19. All publications were screened for metabolite biomarkers reflecting COVID-19 severity. After screening and exclusions (due to non-relevant methodological approaches or inaccessible full-texts), 20 articles were included.
One selected study reported five plasma metabolites – aspartate, malate, guanosine monophosphate (GMP), D-xylulose-5-phosphate, and carbamoyl phosphate, were downregulated in severe COVID-19. In addition, it identified changes in amino acid metabolism in COVID-19 individuals affecting glutamine, arginine, branched amino acids, and their derivatives.
These biological signatures were sustained in patients with mild/moderate and severe COVID-19. Moreover, glutamate remained dysregulated in longitudinal analyses across COVID-19 waves. Some studies also observed alterations in tryptophan metabolism; further, an association between immunosuppressive tryptophan metabolites (anthranilic acid and kynurenine) and COVID-19 severity has also been reported.
Such elevations were also observed in long COVID or cancer patients. Additionally, numerous studies have focused on alterations in carbohydrate and energy metabolism, purine metabolism, tricarboxylic acid cycle, polyamines, and nicotinamide metabolites and identified significant changes in glycerophospholipids in severe COVID-19 patients.
For example, sphingosine-1-phosphate levels increase significantly during recovery, while higher creatine and acetylated polyamine levels may indicate renal dysfunction in critical patients. Furthermore, a meta-analysis of COVID-19 metabolomes identified cholesterol, tyrosine, bilirubin, L-phenylalanine, and D-mannose as the major biomarkers, suggesting that disease severity was associated with changes in pathways involving the indicated metabolites.
Untargeted metabolomic analysis of blood/saliva samples revealed two molecules (L-proline betaine and glycolithocholic acid 3-sulfate) predicting severity. Another study showed that salivary concentrations of valine, proline, tyrosine, phenylalanine, and leucine could differentiate patients with mild and severe COVID-19.
Analysis of nasopharyngeal swabs from patients with mild COVID-19 identified methionine sulfoxide, carnosine, and beta-hydroxybutyric acid, compared to those infected with other respiratory viruses. In addition, a study revealed elevated levels of urea, lactate, and cyclohexane carboxylic acid but reduced levels of D-cellobiose, 1-pentadecanol, propanoic acid, and monomethyl succinate in fecal specimens from severe COVID-19 patients relative to those with mild illness.
Increased L-cytosine levels correlate with SARS-CoV-2 infection and might be deemed a relevant diagnostic biomarker, albeit the mechanistic basis remains unknown. Given the lack of large-scale validation, it remains elusive which specimen type (plasma, saliva, urine, feces, etc.) would be optimal for COVID-19 detection.
Mechanistic COVID-19 biomarkers
COVID-19 amplifies the degradation of glutamine and elevates glutamate levels. Intriguingly, a study revealed that comorbidity-related glutamine deficiency predisposes individuals to severe COVID-19. Polyamines are critical to maintaining cellular homeostasis. Studies have uncovered an overabundance of polyamines and L-ornithine in COVID-19 patient sera.
Further, 3-hydroxybutyric acid (3HB) was found to be elevated during COVID-19. 3HB has immunostimulatory properties and inhibits the inflammasome. In mouse models, 3HB augmented the function of cluster of differentiation 4-positive (CD4+) T cells and decreased the mortality of infected mice. Therefore, it might serve as a metabolite attenuating COVID-19.
Concluding remarks
Taken together, metabolomics has been continuously refined over the past decades to allow for more coverage of the metabolome. The authors reported a catalog of COVID-19-relevant biomarkers. Notwithstanding, standardization of the metabolomic workflow needs to be addressed. Future studies should determine which biomarkers, whether alone or in combination, best reflect infection and COVID-19 severity.