G protein-coupled receptors (GPCRs) remain the subject of intense research for the pharmaceutical and medical fields as they play a key role in transmitting cellular responses to external stimuli. They have been the target of nearly 30% to 50% commercially available pharmaceuticals, highlighting their significance.
Fluorescence resonance energy transfer (FRET), fluorescent ligands, and radioactivity are some of the existing assay techniques used in the analysis of ligands binding to GPCRs. An appropriate substrate needs to be added in the case of bioluminescence resonance energy transfer (BRET) in order to generate the energy capable of non-radiatively exciting fluorescently labeled ligands as well as donor light emission.
In a suitable transfected cell line, it is possible to express the donor luciferase enzyme on the N-terminus of the receptor of interest. The sensitivity of BRET is theoretically better than assays based on FRET. However, identifying an appropriate luciferase enzyme compatible to such kind of N-terminal fusion is practically very challenging, affecting the production of a BRET assay to perform ligand binding studies.
NanoLuc® (Nluc), a genetically engineered luciferase developed by Promega researchers, can be transferred to the cell membranes efficiently as it is readily expressed on the GPCR’s N-terminus when compared to other luciferase-GPCR fusion constructs. Nluc is derived from a naturally available luciferase extracted from deep-sea shrimp. It has relatively low molecular weight (19kDa) and exhibits high stability at temperatures up to 55°C.
Assay Principle
The example illustrated here is a GPCR expression in live cells and Nluc luciferase is on the GPCR’s N-terminus. A fluorophore having an excitation spectrum that overlaps the Nluc’s emission spectrum is used to label a ligand of this receptor. The next step is introducing the ligand to the live cells and adding the Nluc substrate (furimazine).
The detection of the blue emission of Nluc indicates the lack of binding between the ligand and the receptor. The detection of red fluorescence, which is caused by the excitation of the fluorescent label of the receptor by the transfer of resonance energy from the Nluc, indicates the specific binding of the ligand to the receptor. However, the red fluorescence decreases in a concentration-dependent manner due to competitive binding of unlabeled ligand (Figure 1).
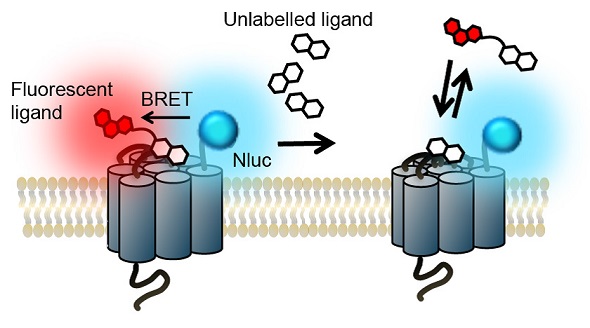
Figure 1. The NanoBRETTM binding assay principle. Image credits: BMG Labtech.
Materials and Methods
The online methods of Stoddart et al. describe the materials and methods in detail.
Stable cell line generation
Promega provided the HEK293 cell line that expresses ß2 adrenergic receptor (ß2AR)
Fluorescently labeled ligands
Promega synthesized alprenolol-TAMRA and CellAura provided propranolol-BY630. Tocris provided CGP 12177, ICI 118551, propranolol, and isoprenaline, and Sigma supplied alprenolol.
Nluc and Rluc8 emission spectra
After 24 hour transfection, cells that express either Nluc-ß2AR or Rluc8-ß2AR were subjected to incubation at 37°C for 180 minutes in OptiMEM in 96 well plates, in the absence of phenol red (Gibco). This was followed by the addition of 10μM luciferase substrates furimazine (Nluc) or coelenterazine h (Rluc8) to the wells.
BRET ß2AR-ligand binding assays
The assessment of HEK293 cells that were transfected with Nluc-ß2AR was carried out in 96-well plates. The saturation binding experiments with alprenolol-TAMRA were then performed by adding serially diluted alprenolol-TAMRA to the wells, with or without 10μM alprenolol, and incubating the plate at room temperature for 120 minutes. The addition of the furimazine substrate brought the final concentration to 10μM. The next step was measuring BRET at room temperature. The competition binding experiments involved incubating the Nluc-ß2AR stably transfected HEK293 cells with the competing ligand diluted to the required concentration in HEPES-buffered saline solution, and 10nM propranolol-BY630 at 37°C for 1 hour.
Instrument Settings
Nluc and Rluc8 emission spectra
A CLARIOstar plate reader was used to determine the emission spectra with the monochromator’s luminescence scanning option: 1nm resolution; 20nm bandwidth; integration time: 500ms with gain: 3000.
BRET ß2AR Ligand Binding Assays
The saturation binding experiments with alprenolol- TAMRA were performed by operating the CLARIOstar plate reader in endpoint mode. Emission filters used were >610nm (longpass) and 450±40nm (80nm bandpass). For each well (well multichromatics), the wavelengths were sequentially read at room temperature, with 500ms as integration time and gain of 3000 for both channels. The saturation binding experiments and the competition binding experiments with propranolol-BY630 were carried out using the PHERAstar FS in endpoint mode and NanoBRETTM module (460nm (80nm bandpass) and >610nm (longpass)), measuring both wavelengths concurrently at room temperature using 1000ms integration time (>610nm gain, 3000; 460nm gain, 2800). The >610nm emission was divided by the 460nm emission to obtain the raw BRET ratio. The use of the term “raw BRET ratio” is due to lack of subtraction of the background ratio.
Results and Discussion
A luminescent signal was observed in initial experiments for sets of HEK293 cells that expressed the ß2AR N-terminally labeled with either Nluc or Rluc8 upon adding their respective substrates. Figure 2 shows the Nluc signal maximum at 460nm in comparison with 480nm for Rluc8. Nevertheless, Nluc had a relative signal strength 70 times higher than Rluc8.
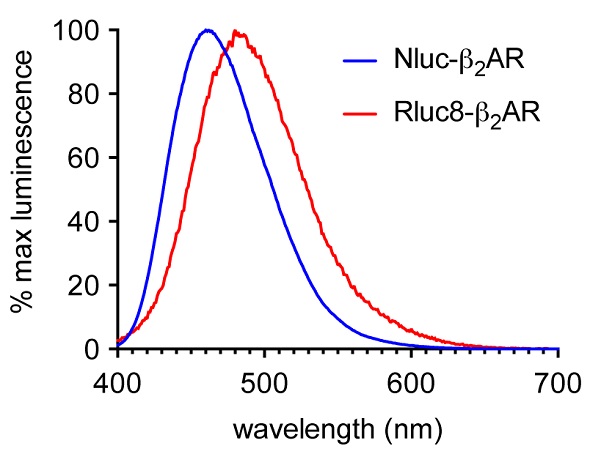
Figure 2. Normalized emission spectra of HEK293 cells expressing ß2AR N-terminally labeled with either Nluc (blue) or Rluc8 (red) using the CLARIOstar. Data previously published in Stoddart et al. Image credits: BMG Labtech.
Nluc’s emission spectrum was maximum at 460nm, but the appreciable intensity of the spectrum was roughly 600nm (Figure 2), thus enabling different fluorescent labels to be used on the desired agonist and antagonist GPCR binding partners. TAMRA (Ex 565, Em 580) was used to label the ß2AR antagonist alprenolol for HEK293 cells that expressed Nluc-ß2AR.
Competitive binding of 10μM unlabeled alprenolol could completely prevent the specific ligand binding observed in Figure 3. The next step was assessing a second fluorescent ligand, propranolol-BY630. An equilibrium binding constant (KD) was estimated to be 18.9 ± 4.1nM from the saturation binding experiments using this labeled antagonist that binds to the receptor.
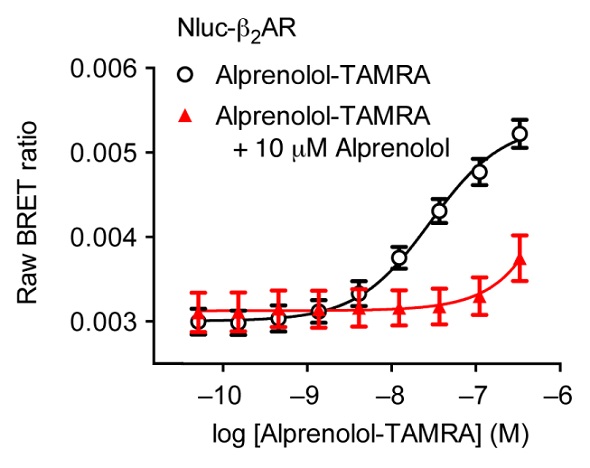
Figure 3. Saturation binding experiment using HEK293 cells expressing Nluc-ß2AR with increasing concentrations of alprenolol-TAMRA (black) and with increasing concentrations of alprenolol-TAMRA in the presence of a high concentration of unlabelled alprenolol (red). Data previously published in Stoddart et al. Image credits: BMG Labtech.
When the concentrations of the agonist isoprenaline and the unlabeled ß2AR antagonists propranolol, ICI 119551 and CGP 12177, were increased, there was a possible decrease in the BRET signal produced as a result of interactions between the receptor and propranolol-BY630 (Figure 4). It is possible to calculate pKi values from these experiments, using the IC50 values of the three antagonists: propranolol (8.13 ± 0.05), ICI 119551 (8.04 ± 0.04) and CGP 12177 (8.32 ± 0.03). Moreover, the resulting values were in good agreement with the values obtained from the radio ligand binding experiments, corroborating the applicability of this assay system for use in drug discovery and profiling.
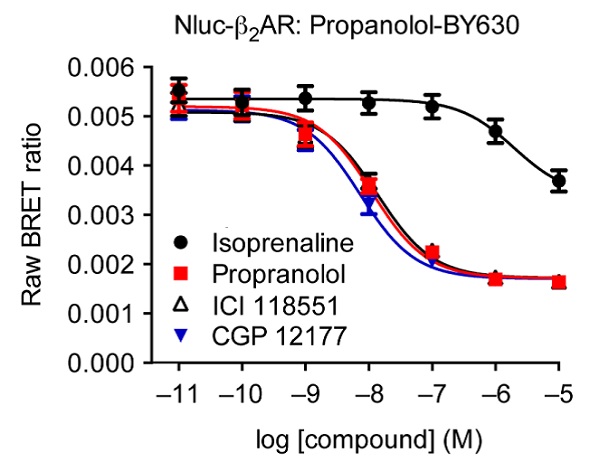
Figure 4. Competitive binding experiments of propranolol-BY630 with increasing concentrations of known labeled ß2AR ligands. Data previously published in Stoddart et al. Image credits: BMG Labtech.
Conclusion
From the results presented in this article, it is evident that the NanoBRETTM assay is a feasible alternative to current techniques to determine the binding of ligands to GPCRs. The technique can be used in fundamental research and in the pharmaceutical sector.
References
- Pfleger, K.D.G. & Eidne, K.A. Nat. Methods 3, 165–174 (2006).
- Hall, M.P. et al. ACS Chem. Biol. 7, 1848–1857 (2012).
- Stoddart, L. et al. Nat. Methods 12, 661-663 (2015).
- Baker, J.G. Br. J. Pharmacol. 144, 317–322 (2005).
Acknowledgements
Produced from materials authored by Delphine Jaga1 and Franka Maurer2
1Cisbio Bioassays, France
2BMG LABTECH, Germany
About BMG Labtech

BMG LABTECH has been committed to producing microplate readers for more than twenty years. By focusing on the needs of the scientific community, the company’s innovative microplate readers have earned the company the reputation of being a technology leader in the field.
BMG LABTECH has developed a wide range of dedicated and multi-mode microplate readers for life sciences applications and high-throughput screening.
All BMG LABTECH microplate readers are "Made in Germany" and are conceived, developed, assembled, and tested entirely at our headquarters in Germany.
Since our establishment in Offenburg, Germany in 1989, BMG LABTECH has expanded to offer a worldwide sales and support network with offices in the USA, UK, Australia, Japan and France. Our subsidiaries, regional offices and distributors are committed to bringing you innovative microplate reader technology with the quality and reliability you expect from a German company.
Our staff includes engineers and scientists from the fields of biology, biochemistry, analytical chemistry, and physics.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.