A routine analysis performed in life science laboratories is the measurement of protein concentration in liquid samples. Accurate measurement is often essential for subsequent analyses such as western blots or protein characterization.
Absorbance-based techniques are cheap, easy to handle, and well-established. They can be carried out in microplates, requiring low reagent volumes and enabling simultaneous processing of high sample numbers.
Spectrometer-based BMG LABTECH readers acquire absorbance at discrete wavelengths or absorbance spectra between 220 and 1000 nm within a second per well, so these assays can be easily read. This article discusses the most common protein quantification methods that are absorbance at 280 nm, Bradford, Bicin-Choninic Acid (BCA), and Lowry assays.
Materials and methods
- BMG LABTECH’s SPECTROstar® Nano, FLUOstar® Omega, PHERAstar® FSX and CLARIOstar® plate readers with rapid and precise default settings
- Clear 96-well plates from Greiner
- Reagents from Sigma Aldrich
- The protein standard Bovine Serum Albumin (BSA) dissolved in ddH2O to a stock of 2 mg/ml
Results and discussion
Absorbance at 280 nm
The UV light is absorbed at 280 nm wavelength by the aromatic residues of tyrosine and tryptophan amino acids (Figure 1A), reflecting the protein concentration. This quantification method is quick and simple as it is not necessary to add further reagents. The concentration range of 125-1000 µg/ml enabled reliable detection of BSA using the BMG LABTECH LVis plate and a volume down to 3 µl (Figure 1B).
- 3 µl (BSA in ddH2O) on LVis plate in duplicates
- Absorbance spectrum 240-400 nm, analysis of A280
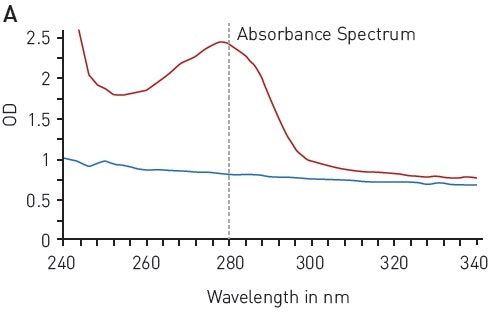
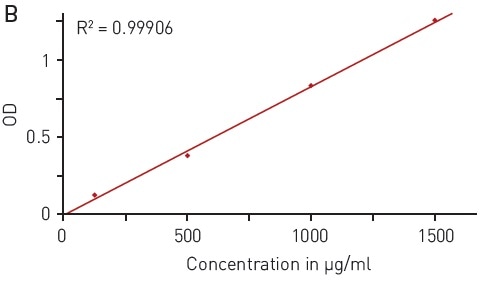
Figure 1. Total protein quantitation by absorbance at 280 nm (A280) A) absorbance spectrum of water (blue) and BSA (2 mg/ml in ddH2O, red) B) Protein standard curve of BSA. Image credits: BMG Labtech.
Bradford assay
The Bradford assay makes use of an absorbance shift of Coomassie Brilliant blue G-250 from 460 nm, in its free state, to 595 nm, if complexed with proteins (Figure 2A). The absorbance of protein samples that are not known is related to samples containing defined protein concentrations (for instance, BSA) that are measured in parallel. A linear signal is provided by the assay in the range of 62.5-1000 µg/ml BSA (Figure 2B).
- 5 µl sample + 250 µl Bradford solution - Shake for 30 seconds
- Incubation in the dark for 5 to 45 minutes - Measure absorbance 400-700 nm (analysis at 595 nm)
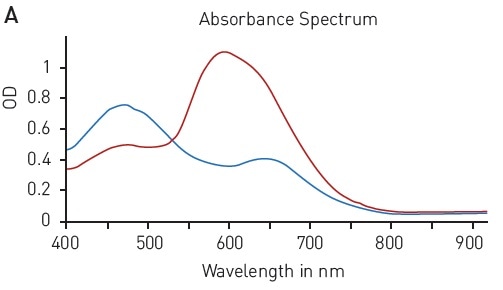
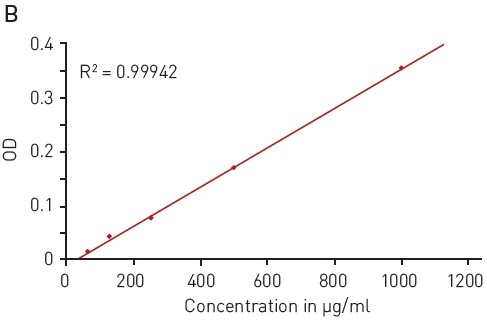
Figure 2. Bradford protein quantification assay. A) Absorbance spectrum of Coomassie Brilliant Blue G 250 (without protein – blue, in presence of BSA – red) B) Protein standard curve of BSA. Image credits: BMG Labtech.
BCA assay (bicinchoninic acid)
The ability to reduce Cu (II) sulfate to Cu+ by peptide bonds is the basis of the BCA assay. This reduction forms complexes with two BCA molecules, providing a chromophore with an absorbance maximum at 562 nm (Figure 3A).
In this assay, the quantification of an unknown protein sample is related to a calibration curve acquired with samples of known protein concentration. With the BCA assay, a broad BSA concentration range of 15.63 to 1500 µg/ml can be quantified using a hyperbola fit (Figure 3B).
- 240 µl working reagent + 30 µl sample
- Incubate at 37 °C for 30 minutes - Measure absorbance 450-750 nm (analysis at 562 nm)
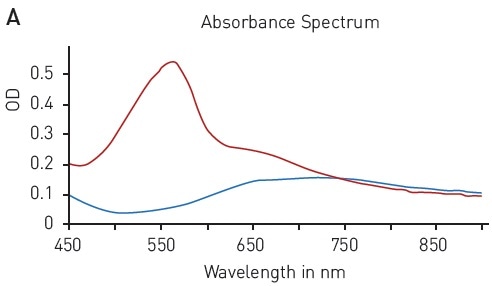
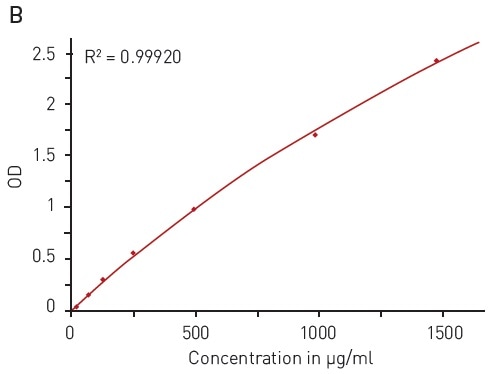
Figure 3. BCA protein quantification assay. A) Absorbance spectrum of bicinchoninic acid (native – blue, BCA-Cu+-complex in presence of BSA - red) B) Protein standard curve of BSA. Image credits: BMG Labtech.
Lowry assay
Likewise, the modified Lowry assay is based on the ability of peptide bonds to reduce copper (II) sulfate. The resulting Cu+ protein complex reacts with Folin–Ciocalteu reagent to form a chromophore with a wide absorption spectrum of 500-800 nm (Figure 4A).
This quantification assay needs a calibration curve to be measured in parallel. With the modified Lowry assay, BSA solution can be reliably detected between 15.63 and 1000 µg/ml using a hyperbola fit (Figure 4B).
- 100 μl sample + 100 μl Lowry reagent - Incubate 20 minutes
- Add 50 μl Folin & Ciocalteu’s Phenol Regent and mix
- Incubate for 30 minutes - Measure 500-800 nm (analysis at 710 nm)
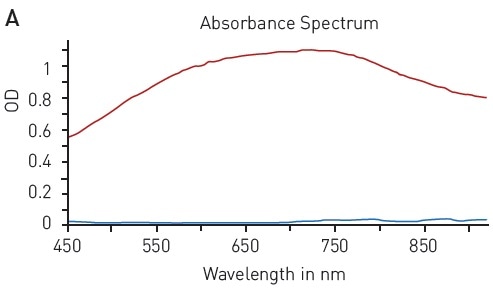
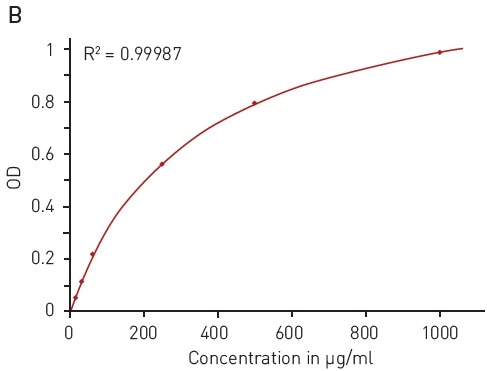
Figure 4. Modified Lowry protein quantification assay. A) Absorbance spectrum of Lowry reaction (without protein - blue, in presence of BSA - red) B) Protein standard curve of BSA. Image credits: BMG Labtech.
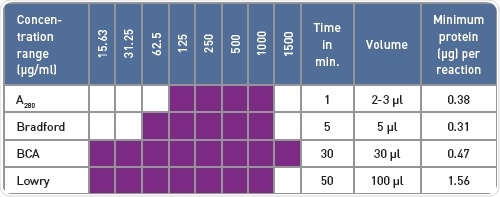
Table 1. Comparison of protein quantification methods
Conclusion
BMG LABTECH readers easily detected the standard absorbance-based protein quantification assays. The protein quantification techniques differ in terms of the volume required, covered concentration range, and the time of incubation and preparation (Table 1), but the appropriate quantification relies on other factors such as detergents present in the buffer that interfere differently with the assays.
Acknowledgements
Produced from materials originally authored by Andrea Krumm from BMG LABTECH GmbH, Ortenberg, Germany.
References
- Bradford, M.M. (1976) Anal, Biochem., 72,248-254.
- Lowry, O.H., et al. (1951) J. Biol. Chem., 193, 265-275.
- Smith PK. et al. (1985) Anal. Biochem. ,150(1), 76-85.
About BMG Labtech

BMG LABTECH has been committed to producing microplate readers for more than twenty years. By focusing on the needs of the scientific community, the company’s innovative microplate readers have earned the company the reputation of being a technology leader in the field.
BMG LABTECH has developed a wide range of dedicated and multi-mode microplate readers for life sciences applications and high-throughput screening.
All BMG LABTECH microplate readers are "Made in Germany" and are conceived, developed, assembled, and tested entirely at our headquarters in Germany.
Since our establishment in Offenburg, Germany in 1989, BMG LABTECH has expanded to offer a worldwide sales and support network with offices in the USA, UK, Australia, Japan and France. Our subsidiaries, regional offices and distributors are committed to bringing you innovative microplate reader technology with the quality and reliability you expect from a German company.
Our staff includes engineers and scientists from the fields of biology, biochemistry, analytical chemistry, and physics.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.