Overview
Using standard PCR systems, users need to set a predefined number of cycles according to the assay and the input amount. This necessitates the quantification of input material as well as splitting of different input samples among various PCR runs.
FFPE samples, which have different levels of genomic integrity, complicate standard PCR settings where the input quantity no longer correlates with the quantity of amplifiable fragments. The need to process samples of similar integrity and input quantities at the same time necessitates several thermocycler runs and workflows capable of incorporating pre- and post-PCR quality control steps on each and every sample individually.

Figure 1. iconPCR, the world’s first real-time thermocycler with 96 individually controlled wells. Image Credit: n6
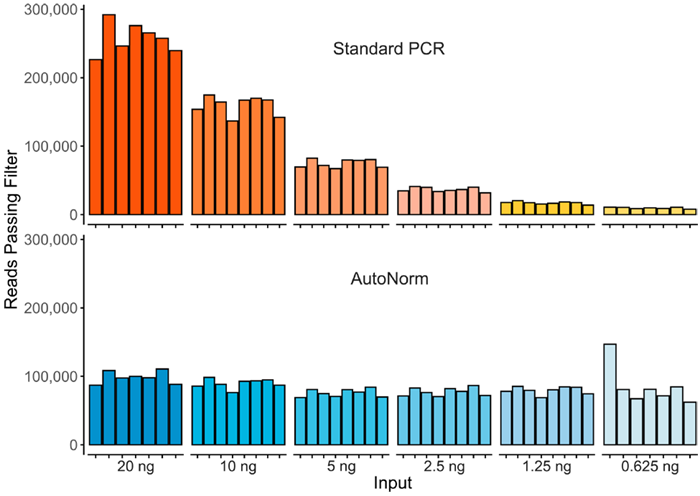
Figure 2. Here, we show the significant yield variance when using a single PCR instrument with a fixed number of PCR cycles compared to iconPCR with AutoNorm, where each sample is amplified to similar levels. Image Credit: n6
iconPCR, built by n6, constitutes a dramatic shift in PCR technology. Its central innovation arises from AutoNorm™, wherein every individual PCR reaction is tracked and concluded in real-time according to fluorescence thresholds.
This eliminates the guesswork inherent to fixed-cycle PCR and ensures that every library is amplified in the best way. iconPCR facilitates new single-cell workflows, making it possible for various sample types and inputs to run at the same time, increasing efficiency and enabling security against incorrect cycling conditions.
To assess iconPCR’s performance, n6 prepared whole-genome sequencing libraries from FFPE and high molecular weight control samples. The use of AutoNorm enabled simultaneous processing of samples over several DNA integrities and input ranges.
Experimental design
Libraries were produced using Standard or iconPCR with AutoNorm with one, 10, and 100 ng of DNA from FFPE (colon, liver) and control (NA12878) samples.
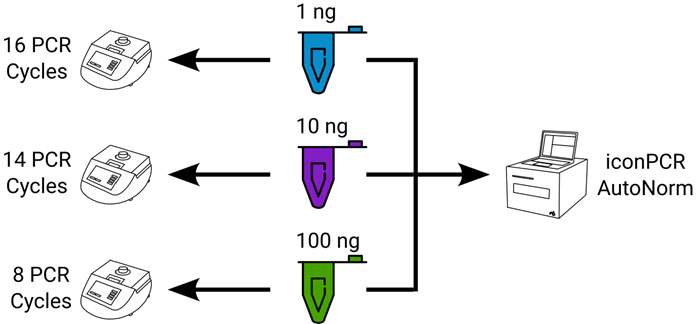
Figure 3. Experimental design showing sample distribution across workflows. For each tissue tested, 1 ng, 10 ng, or 100 ng of total DNA was used as input. For standard PCR, each input amount was used with a fixed number of PCR cycles, per manufacturer recommendations, requiring three separate thermocycler runs. For iconPCR with AutoNorm, all samples were run concurrently. Image Credit: n6
Optimized workflow
Standard PCR requires three separate thermocycler runs of 16, 14, and eight cycles, linked to inputs of one ng, 10 ng, and 100 ng. However, with iconPCR, each sample, regardless of input amount, was run on the iconPCR simultaneously.
With AutoNorm, cycle numbers were calculated dynamically, with each of the wells stopping when fluorescence reached a target value of 3000 relative fluorescence units (RFU).
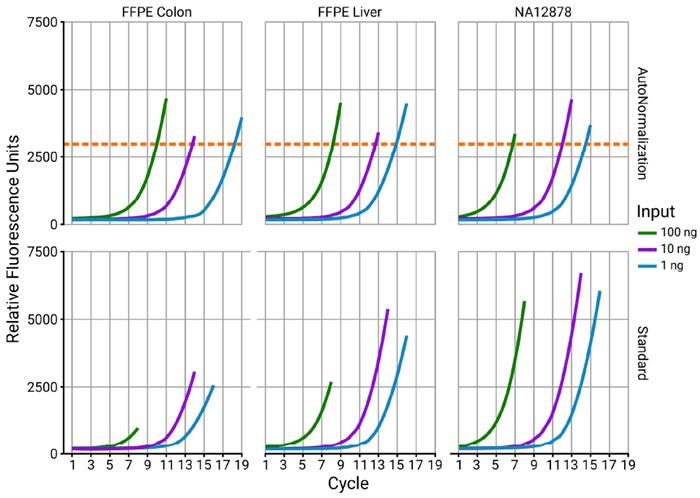
Figure 4. Proper amplification of all samples in a single run with AutoNorm. Standard PCR showed a high degree of variability in the endpoint fluorescence between different inputs within a tissue as well as between different tissues with the same inputs. However, using AutoNorm on iconPCR, samples cycle until they reach the 3000 RFU threshold (orange dotted line), ensuring that samples receive the proper amount of cycles. Image Credit: n6
Table 1. AutoNorm dynamically controls cycle numbers. The table shows the stop cycle for each sample in the study. Using AutoNorm allows each sample to stop at a different number of cycles, ensuring that each sample, if properly amplified, all within a single PCR run. Source: n6
|
Standard PCR |
AutoNormalization |
| Input |
All Samples |
FFPE Colon |
FFPE Liver |
NA12878 |
| 1 ng |
16 |
19 |
16 |
15 |
| 10 ng |
14 |
14 |
13 |
13 |
| 100 ng |
8 |
11 |
9 |
7 |
Data quality
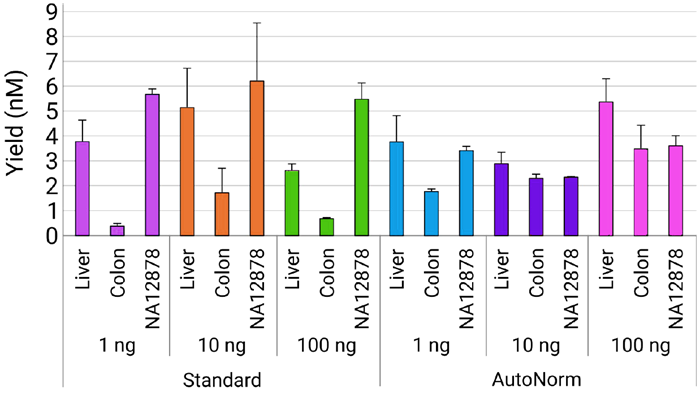
Figure 5. iconPCR allows for effective normalization of output across input amounts and sample variability. Following library preparation, samples were SPRI cleaned and quantified via Tapestation. iconPCR samples showed much less variability in concentration than those amplified using standard PCR. Image Credit: n6
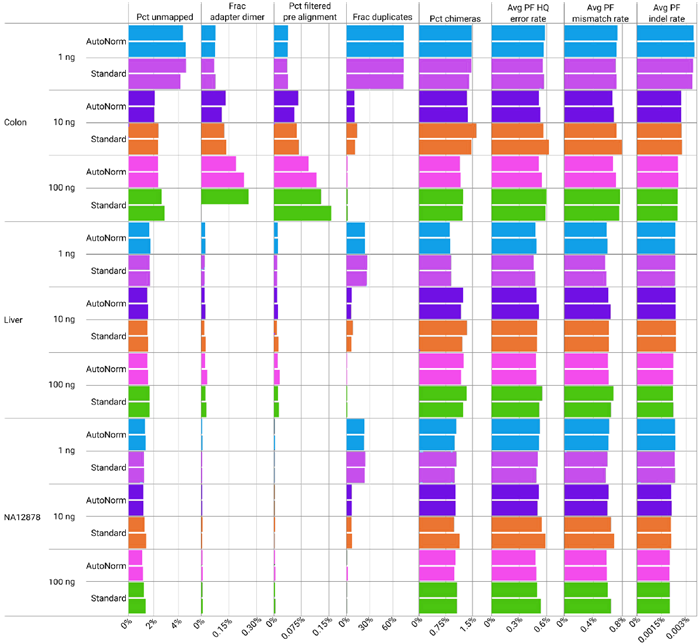
Figure 6. Alteration of cycle numbers has no effect on data quality. All samples, regardless of preparation method, showed similar quality metrics post sequencing. Image Credit: n6
Advantages of iconPCR. Source: n6
| Conventional PCR |
iconPCR (with AutoNorm) |
| Fixed cycle count (e.g., 30 cycles) |
Real-time fluorescence monitoring |
| One-size-fits-all amplification |
Per-well cycle control based on signal, not guesswork |
| Under/over-amplification common |
Optimal amplification per sample |
| High chimera rates |
Reduced chimera formation |
| Variable library quality |
Uniform library quality across all wells |
| Manual quant and normalization required |
Automated normalization (no post-PCR quant) |
| More hands-on time |
40–60 % reduction in hands-on time |
| Increased reagent waste |
Lower reagent waste, fewer failed libraries |
| Extra QC and rerun costs |
Faster turnaround, minimal QC/rescue steps |
Conclusion
iconPCR brings new life to library preparation by removing the limitations inherent to conventional PCR workflows. Using its unique per-well AutoNorm mechanism, iconPCR produces streamlined workflows and eliminates challenges that arise from FFPE samples without reducing data quality.
About n6
n6 proudly introduces iconPCR, a pioneering advancement in the genomics field with the world’s first real-time thermocycler with 96 individually controlled wells. This breakthrough technology promises to revolutionize DNA amplification and sequencing by offering unmatched simplicity and flexibility, setting a new standard for genomic research and diagnostics.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.