The low-pass whole genome sequencing technique involves sequencing each genome base several times. It is increasingly popular in agricultural studies, as agricultural genomes typically have a broader range of species and are significantly larger than human genomes.
The average coverage depth is less than five times, which enables the sequencing of many genomes simultaneously, and the low coverage depth also reduces sequencing costs. This approach identifies known SNP markers and novel content such as repeats, non-coding sequences, and transposable elements.
However, reductions in sequencing costs have shifted the majority of expenses to library preparation, such as DNA quantification, purification, and balanced pooling, which require significant time, manpower, and resources. A key problem is maintaining sample representation owing to dropout or over-sequencing.
Using the iconPCR system, n6 presents enhancements in low-pass whole genome sequencing. This approach enables balanced, pooled libraries at a significantly lower cost by eliminating more than 66 % of the manual steps in library preparation.
AutoNormalization allows variable DNA input amounts, sample-specific amplification, consistent yields, and single-tube SPRI purification. This diminishes the need for manual pooling and liquid-handling robots.
The system uses a 96-well real-time thermocycler; each well is controlled independently. Libraries are amplified to the target level, and cycling stops when the needed concentration is attained. The 96-well plate is mixed into a single multiplex pool and purified in one step, minimizing labor-intensive processes such as purification, quantification, and manual mixing.
Simplifying library prep to match ultra-high throughput sequencing
This research demonstrates the elimination, or drastic reduction, of individual library prep steps with the implementation of iconPCR. A variety of input masses can be used without labor-intensive processes to oversee variability. Examination of the amplification plots can be used to detect sub-optimal amps, dropouts, and reagent quality problems.
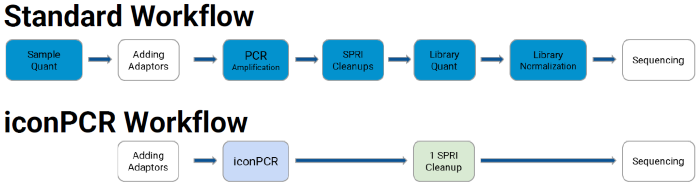
Simplified workflow using standard reagents and consumables. Image Credit: n6
Workflow enhancements:
- Eliminating input mass normalization
- Use of existing consumables and reagents
- Significant reduction in both pipetting steps and tips
- Lower error opportunity
- Single plate amplification vs. three different runs
- >90 % drop in hands-on-time for SPRI
- AutoNormalized libraries that are ready for sequencing
96 - plex droplet-emulsion PCR (dePCR) preparation of WGS libraries study goals:
To examine workflow optimization and library preparation efficiency, a 96-reaction study was designed to examine significant reductions in the hands-on-time and manipulation steps using liquid handling robotics.
In this research, DNA from the Genome in a Bottle sample HG006 (NA24694) was prepared in a dilution series from 10 ng to 120 ng input amounts in a 96-well plate, using eight replicates per concentration.
A control plate was prepared and processed using the standard amplification workflow. At the same time, replicate plates were processed via the iconPCR system to detect peak Auto-Normalization settings.
Control Samples: Input vs. # PCR Cycles. Source: n6
| 8 PCR Cycles |
7 PCR Cycles |
6 PCR Cycles |
| 10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
| 10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
| 10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
| 10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
| 10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
| 10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
| 10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
| 10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
| 10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
Control plate setup: The control plate was divided by the input quantity and then amplified in three different thermocycler cycles, allowing for diverse levels of amplification.
After amplification, samples were assembled in a 96-well plate to be processed on a Hamilton liquid handling robot.
IconPCR plate setup: Using a Determination run, the peak thresholds were chosen for the three AutoNorm approaches: Slope, Target Fluorescence (FL) and xBaseline. Three plates were amplified using the various AutoNorm approaches as options for both variable yields and AN levels. The Target FL approach was selected for both sequencing and comparison to the Control.
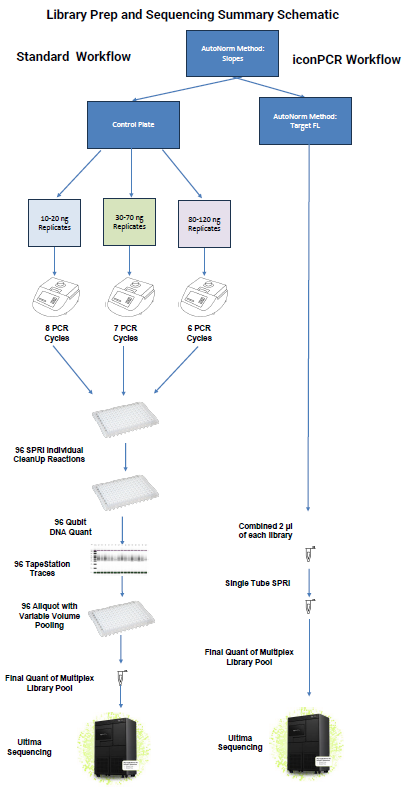
Image Credit: n6
Simplification lowers pipetting error opportunity: iconPCR can rescue hyper-variable challenges
Sample Dropouts: The Control and iconPCR stamped plates both demonstrated a high level of variability in the 30-ng replicate column, suggesting a technical pipetting error. The Control plate recorded four replicate dropouts in the 30 ng column alongside four random dropouts on the plate. The iconPCR plate demonstrated elevated levels of variability, but AutoNorm rescued three of four dropouts from Column 3 and left no random dropouts on the plate.
Control Workflow. Source: n6
|
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
| A |
10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
| B |
10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
| C |
10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
| D |
10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
| E |
10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
| F |
10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
| G |
10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
| H |
10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
iconPCR. Source: n6
|
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
| A |
10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
| B |
10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
| C |
10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
| D |
10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
| E |
10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
| F |
10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
| G |
10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
| H |
10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
Library balancing via manual normalization vs iconPCR AutoNormalization
The Control multiplex library and the AutoNormalized iconPCR pool were both shallow-sequenced via a 2x150 bp sequencing run. The amount of each library was assessed and represented by each barcoded sample index. Average representation and variation (% CV) are showcased for the two workflows.
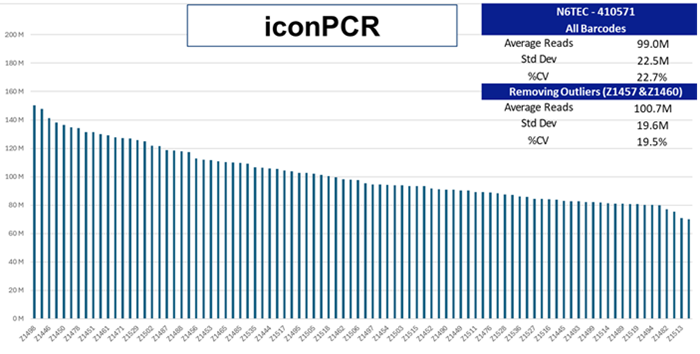
Image Credit: n6
Pooling of the AutoNorm run:
The multiplexed sequencing library was generated by pooling two microliters of each reaction. This approach reflects small variations in the final concentration and is the most coarse method for pooling, reflected in up to a one Ct difference among samples.
Refined balancing can be attained via the iconPCR Pooling using the Fluorescence (FL) approach. This approach uses the ultimate RFU values from each well to adjust each sample’s volume.
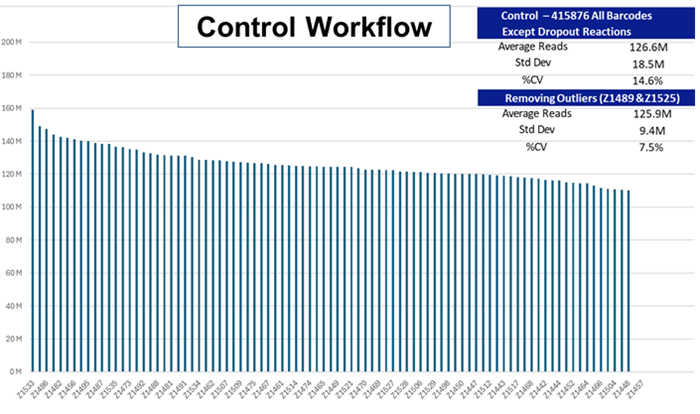
Image Credit: n6
Pooling the control run:
The percent coefficient of variation for the Control run led to a % CV of 7.5 %, demonstrating the elevated precision levels reflecting the extra time, effort, and expense linked to legacy NGS workflows.
Pooling by FL will lead to barcode balancing similar to manual pooling
Pooling by Vol versus pooling by FL (example):
Previous research has shown that using the final RFU value enables more precision in assay types that require greater sequencing requirements for each sample, like WGS. For sequencing-light assays, like RNA-seq, 16S, and scRNA-seq, pooling by volume enables a simplified automation workflow, as each sample is allotted at the same volume.
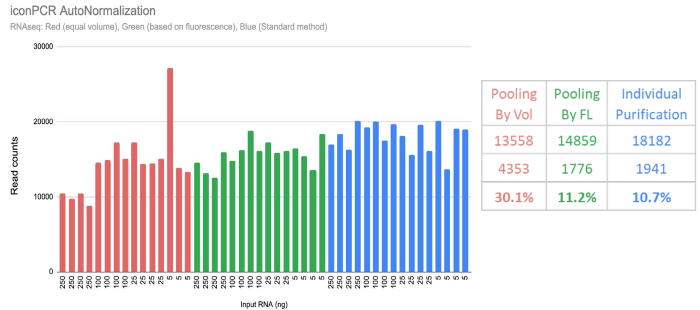
Image Credit: n6
Example of pooling methods:
A 50X fold dilution series was produced for RNA-seq libraries spanning from five to 250 nanograms of input. Libraries were amplified via iconPCR AutoNorm, and the following material was further processed using standard purification, quant, and pooling of each library (standard) alongside the two approaches recommended by n6: Pooling by Vol and Pooling by FL.
Therefore, researchers can select the most economic and efficient sequencing options according to assay type.
Pooling by FL provides equal library balancing as manual method:
Adjusting the pooling volume according to the End-point RFU values, it is possible to attain the same level as the laborious legacy approach. N6 provides an analysis tool to make recommendations for all approaches directly.
Special thank you and authorship credit to the following:
The Ultima Genomics team: Florian Oberstrass, Tyson Clark, Eric Jaschke, Nika Iremadze, Freemont, CA.
n6 iconPCR team: Yann Jouvenot, Pranav Patel, Wes Austin, Chris Streck, Gagneet Kaur, n6, Pleasanton, CA.
About n6
n6 proudly introduces iconPCR, a pioneering advancement in the genomics field with the world’s first real-time thermocycler with 96 individually controlled wells. This breakthrough technology promises to revolutionize DNA amplification and sequencing by offering unmatched simplicity and flexibility, setting a new standard for genomic research and diagnostics.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.