Overview
16S rRNA gene sequencing is crucial for delving deeper into microbial diversity in ecosystems ranging from soil and water to human-associated microbiomes. However, conventional PCR methods, as used in 16S library preparation, often generate considerable bias owing to over- and under-amplification and the generation of chimeric sequences.
Such biases reduce the precision of microbial community profiling by contorting relative abundances and obscuring low-abundance taxa.
iconPCR, built by n6, represents a dramatic shift in PCR technology. Its key innovation is AutoNorm™, which monitors each PCR reaction in real time and stops it once fluorescence thresholds are reached. This removes the uncertainty of fixed-cycle PCR and ensures optimal amplification for every library

Figure 1. iconPCR, the world’s first real-time thermocycler with 96 individually controlled wells. Image Credit: n6
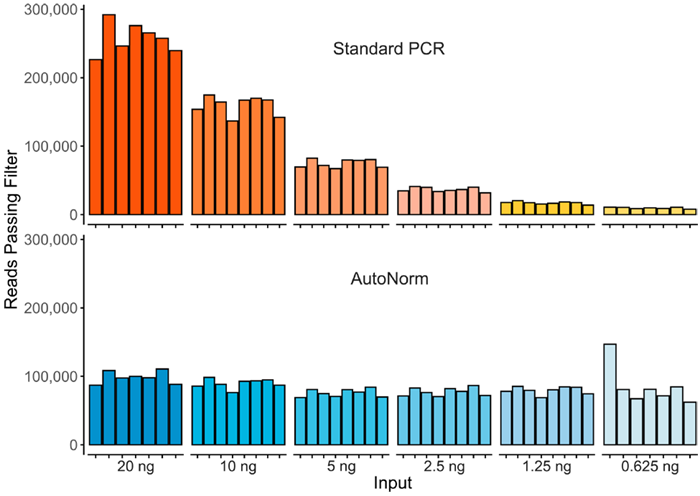
Figure 2. Here we show the significant yield variance when using a single PCR instrument with a fixed number of PCR cycles compared to iconPCR with AutoNorm where each sample is amplified to similar levels. Image Credit: n6
To assess performance, n6 prepared full-length 16S rRNA libraries from soil microbiome samples and sequenced them using the PacBio® Sequel® II platform.
Renowned for its ability to produce accurate, long-read sequencing data, PacBio technology was essential for recording the full-length 16S gene (V1-V9), making high-resolution taxonomic analysis possible. Combining iconPCR and PacBio sequencing resulted in highly reproducible and detailed profiles of complex microbial communities.
This article outlines the comparative study using iconPCR and traditional PCR workflows. The results clearly showed the superiority of iconPCR regarding data quality, taxonomic resolution, and workflow consistency, particularly when coupled with long-read platforms such as PacBio for full-length amplicon sequencing.
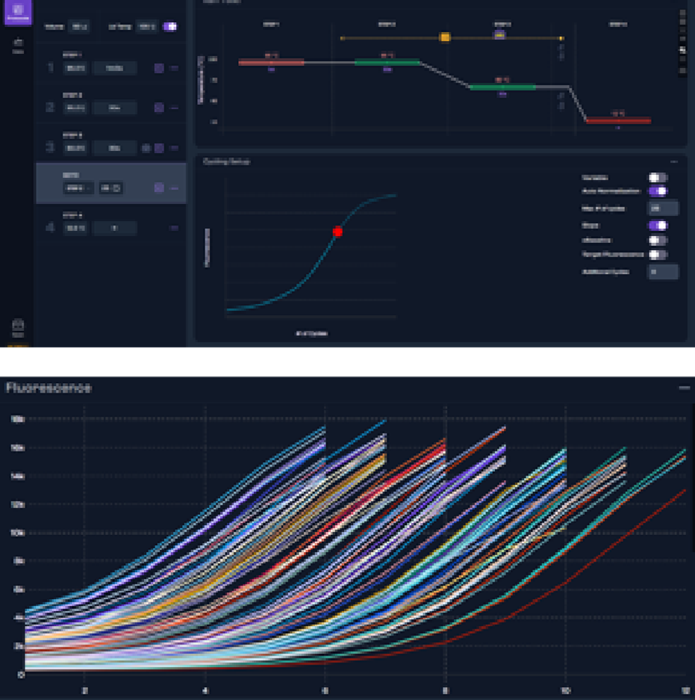
Figure 3a. Graphical view of setting AutoNorm on iconPCR and 3b. resulting in the amplification of each sample to its set threshold. Image Credit: n6
Source: n6
| Conventional PCR |
iconPCR (with AutoNorm) |
| Fixed cycle count (e.g., 30 cycles) |
Real-time fluorescence monitoring |
| One-size-fits-all amplification |
Per-well cycle control based on signal, not guesswork |
| Under/over-amplification is common |
Optimal amplification per sample |
| High chimera rates |
Reduced chimera formation |
| Variable library quality |
Uniform library quality across all wells |
| Manual quant and normalization required |
Automated normalization (no post-PCR quant) |
| More hands-on time |
40–60 % reduction in hands-on time |
| Increased reagent waste |
Lower reagent waste, fewer failed libraries |
| Extra QC and rerun costs |
Faster turnaround, minimal QC/rescue steps |
Experimental design
60 soil samples and 4 ZymoBIOMICS positive controls were divided between two different workflows:
- iconPCR Using AutoNorm Mode (AN)
- Traditional PCR using 30 fixed cycles (FC)
Libraries were sequenced using PacBio Sequel II, targeting full-length 16S (V1-V9). Fluorescence thresholds in iconPCR were set to 2.5x baseline, with each sample terminating on its own at its optimal cycle.
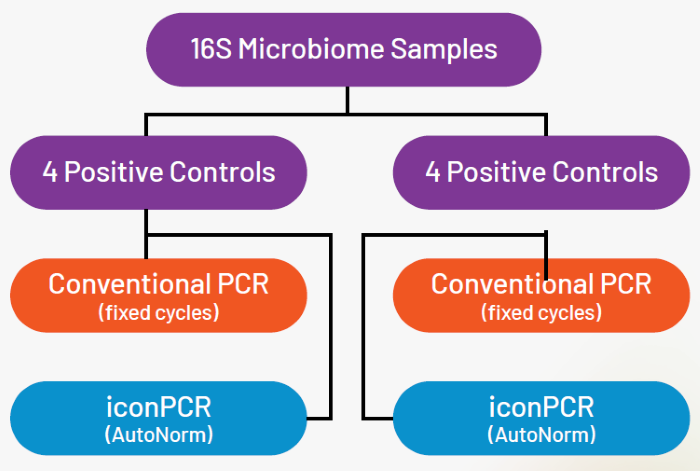
Figure 4. Experimental design showing sample distribution across workflows. Image Credit: n6
Improved data quality
Data was processed using the DADA2 pipeline. iconPCR libraries continuously exceeded traditional PCR in multiple metrics:
- Reduced chimeras in each of the samples
- More trimmed and inferred reads
- Up to 10 x more unique ASVs detected
The comparative analysis between iconPCR and traditional fixed-cycle PCR across 60 soil samples and positive controls shows a consistent and considerable improvement with iconPCR. The two workflows started with the same raw reads (5,400 per sample), but iconPCR maintained more trimmed reads, produced up to 10 × more high-confidence DADA2-inferred reads, and generated zero chimeras across all samples, unlike the fixed-cycle protocol, which frequently yielded significant chimera counts.
Furthermore, iconPCR generated higher chimera-free reads and a considerably higher number of unique amplicon sequence variants (ASVs), signaling more richness and sequencing efficiency. These outcomes prove iconPCR’s superiority in data quality and artifact reduction as well as microbial diversity resolution.
Expanded microbial detection
iconPCR greatly improved species richness and taxonomic depth. Multiple microbial classes, including Chloroflexia, Opitutae, Thermomicrobia, and Caldilinea, were exclusively identified in iconPCR-prepared samples.
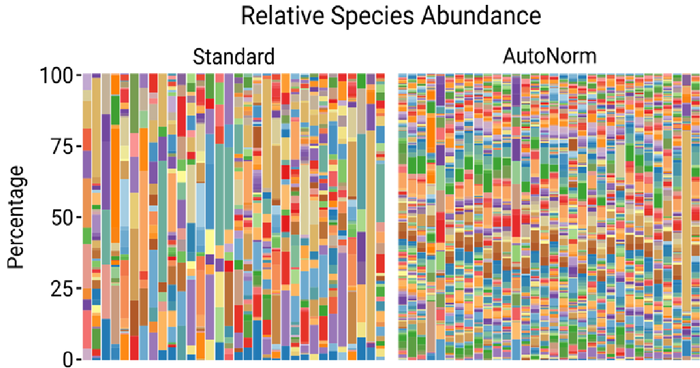
Figure 5. Species-level abundance profiles. Image Credit: n6

Figure 6. Core and unique taxa detected per workflow at the class level. Image Credit: n6
Diversity metrics
Alpha and beta diversity analyses further emphasised the improved performance of iconPCR:
- Alpha Diversity (Chao1, Shannon, Simpson): iconPCR-treated samples showed considerably more diversity (p < 0.001).
- Beta Diversity: PCoA plots using Bray-Curtis distances demonstrated tighter clustering of iconPCR samples, pointing towards more consistency and reproducibility.
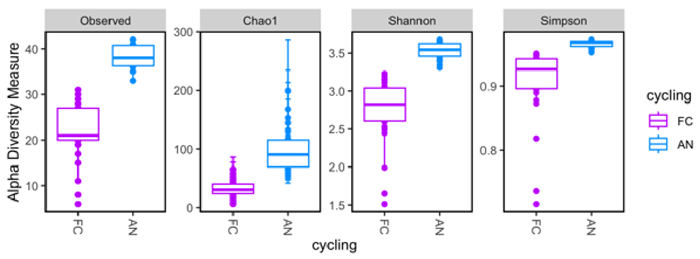
Figure 7. Alpha diversity analysis comparing observed richness and indices. Image Credit: n6
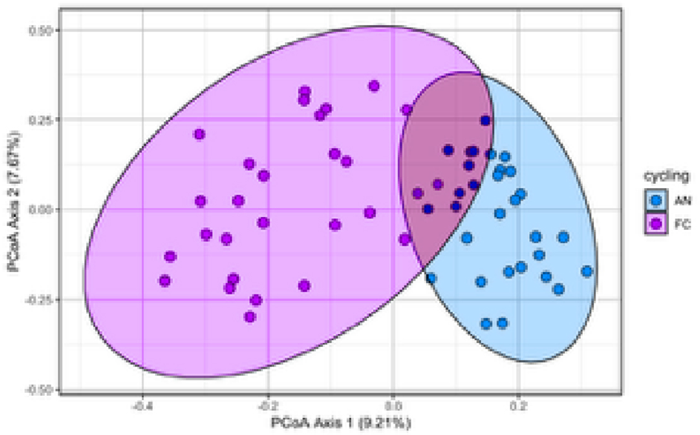
Figure 8. Beta diversity plot (PCoA) showing distinct clustering patterns. Image Credit: n6
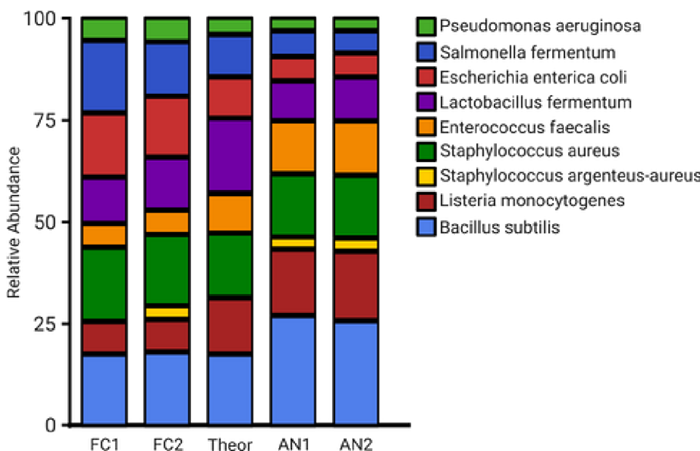
Figure 9. Relative species abundance of positive controls from fixed cycle PCR and iconPCR compared to the theoretical composition. Image Credit: n6
Incorporating iconPCR into your 16S workflow
gDNA extraction
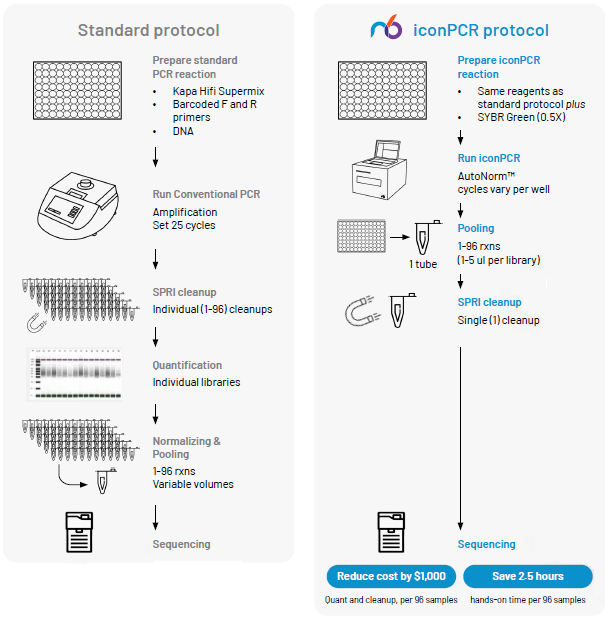
Image Credit: n6
Conclusion
iconPCR redefines 16S library preparation by getting rid of the limitations inherent to conventional PCR workflows. With its special per-well AutoNorm mechanism, iconPCR produces data of higher quality, minimizes artifacts, and enhances taxonomic discovery.
Coupled with long-read sequencing platforms like PacBio, iconPCR can be used to produce a robust, end-to-end workflow for producing high-resolution, chimera-free, full-length 16S profiles. Such benefits make it the optimal solution for microbiome research across environmental, clinical, and industrial use cases, enabling scientists to gather clearer, more reproducible insights into microbial communities.
About n6
n6 proudly introduces iconPCR, a pioneering advancement in the genomics field with the world’s first real-time thermocycler with 96 individually controlled wells. This breakthrough technology promises to revolutionize DNA amplification and sequencing by offering unmatched simplicity and flexibility, setting a new standard for genomic research and diagnostics.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.