Ultima Genomics has transformed highly efficient, production-scale sequencing while reducing the sequencing cost per genome to under $100. This increase in throughput has shifted the overall cost per sample to the library preparation process.
Labor, reagents, and consumables are the main costs and bottlenecks, even when using liquid handling robots. Individual processing of each sample via SPRI cleanup, quantification, QC assessment, and normalized pooling is a significant bottleneck, especially when trying to scale to current sequencing output.
Using iconPCR, this research demonstrates the efficiencies that can be gained by using iconPCR’s AutoNormalization, reducing inefficiencies by more than 95 %.
A dilution series of input DNA ranging between ten and 120 ng was processed and sequenced to show workflow improvements and cost savings while maintaining a high degree of uniformity in representing each sample within a multiplexed sequencing run.
96 - plex droplet-emulsion PCR (dePCR) preparation of WGS libraries
To evaluate workflow optimization and library preparation efficiency, a 96-reaction study was designed to assess significant reductions in the hands-on time and manipulation steps using liquid handling robotics. In this research, DNA from the Genome in a Bottle sample HG006 (NA24694) was prepared in a dilution series from ten to 120 ng input amounts in a 96-well plate, using eight replicates per concentration.
A Control plate was prepared and processed via the standard amplification workflow. At the same time, replicate plates were processed via the iconPCR system to distinguish ideal Auto-Normalization settings.
Control Samples: Input vs. #PCRCycles. Source: n6
| 8 PCR Cycles |
7 PCR Cycles |
6 PCR Cycles |
| 10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
| 10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
| 10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
| 10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
| 10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
| 10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
| 10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
| 10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
| 10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
Control plate setup: The Control plate was divided by input quantity and amplified in three different thermocycler runs to achieve various amplification levels.
After amplification, samples were put together into a single 96-well plate to be processed on a Hamilton liquid handling robot.
IconPCR plate setup: With a determination run, the ideal thresholds were chosen for all three AutoNorm approaches: Slope, Target Fluorescence (FL), and xBaseline. Three plates were amplified with the various AutoNorm approaches to act as options for variable yields and AN levels. The Target FL approach was selected for sequencing and comparison to the Control.
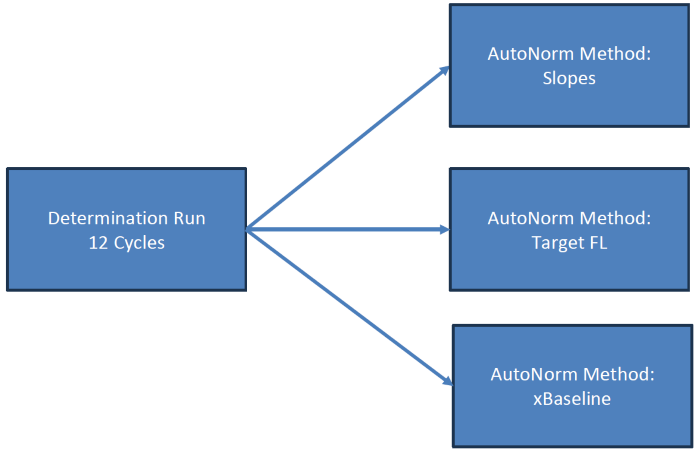
Image Credit: n6
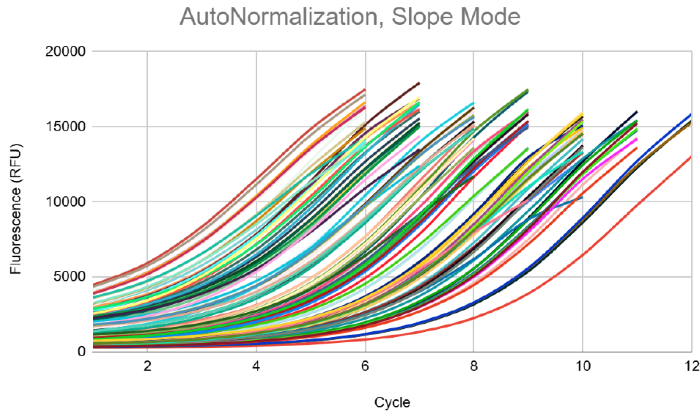
Slope Example Shown Above: Target FL AutoNorm selected for further processing and sequencing. Image Credit: n6
The Target FL plate was chosen for sequencing. A two microliter aliquot from each of the samples was pooled by putting together equal volumes of each reaction into a singular multiplexed pool.
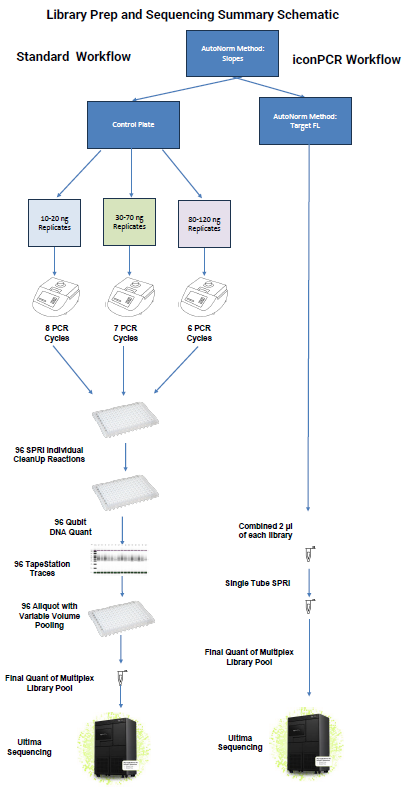
Image Credit: n6
Examining library recovery and sample dropouts
Sample dropouts: The Control workflow captured seven sample dropouts and an elevated degree of variability in the 30 ng input quantity. iconPCR had just one dropout in the 30 ng input column.
Control Workflow. Source: n6
|
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
| A |
10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
| B |
10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
| C |
10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
| D |
10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
| E |
10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
| F |
10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
| G |
10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
| H |
10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
iconPCR. Source: n6
|
1 |
2 |
3 |
4 |
5 |
6 |
7 |
8 |
9 |
10 |
11 |
12 |
| A |
10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
| B |
10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
| C |
10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
| D |
10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
| E |
10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
| F |
10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
| G |
10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
| H |
10 ng |
20 ng |
30 ng |
40 ng |
50 ng |
60 ng |
70 ng |
80 ng |
90 ng |
100 ng |
110 ng |
120 ng |
The column with the 30 ng input demonstrated significant variability in comparison to what remained on the plate in the Control as well as iconPCR plates, perhaps due to a technical error in plate setup. iconPCR rescued three of four of the dropouts seen in the Control plate. Additionally, individual replicates in the 50, 80, and 110 ng inputs generated dropout samples.
Library balancing using manual normalization as opposed to iconPCR AutoNormalization
The Control multiplex library and the AutoNormalized iconPCR pool together were sequenced via a 2x150 bp sequencing run. The amount of each library was assessed and represented by each of the barcoded sample indices. Average representation and variation (% CV) are demonstrated for the two workflows.
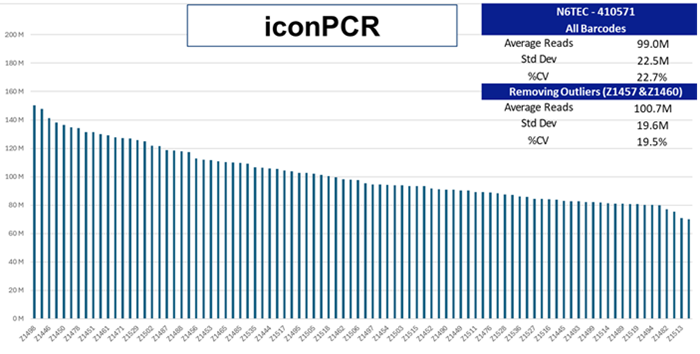
Image Credit: n6
Pooling of the AutoNorm run:
The multiplexed sequencing library was put together by pooling two microliters of each reaction. This approach reflects small differences within the ultimate concentration and is the coarsest way of pooling, reflecting up to one Ct between samples.
Refined balancing may be attained via the iconPCR Pooling by Fluorescence (FL) approach. This approach uses the ultimate RFU values from each well to adjust each sample’s volume.
Pooling by FL will lead to barcode balancing of a similar degree to manual pooling (see below)
Pooling of the control run:
The percent coefficient of variation for the Control run produced a % CV of 7.5 % which demonstrates the elevated precision that reflects the extra time, effort, and costs linked to legacy NGS workflows.
Pooling by Vol as opposed to pooling by FL (example):
Previous research showed that the final RFU value enables more precision for assay types with more significant sequencing needs per sample, like WGS.
For sequencing-light assays, such as RNA-seq, 16S, and scRNA-seq, Pooling by Volume outcomes provide a simple automation workflow as each sample is aliquoted in identical volumes.
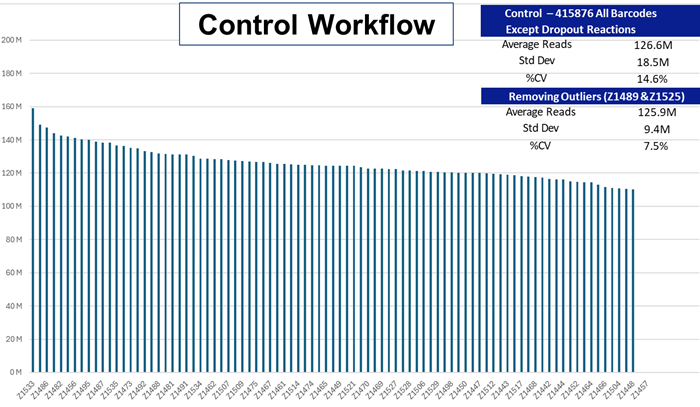
Image Credit: n6
Example of pooling methods:
A 50 x fold dilution series was produced for RNA-seq libraries spanning 250 ng to five nanograms input. Libraries were amplified using iconPCR AutoNorm, and the resultant material was processed further via the standard purification, quant, and pooling of each library (standard) alongside both of n6’s recommended approaches: Pooling by Vol and Pooling by FL.
Scientists can, therefore, review the different approaches to find the one that is most cost-effective and efficient for sequencing according to assay type.
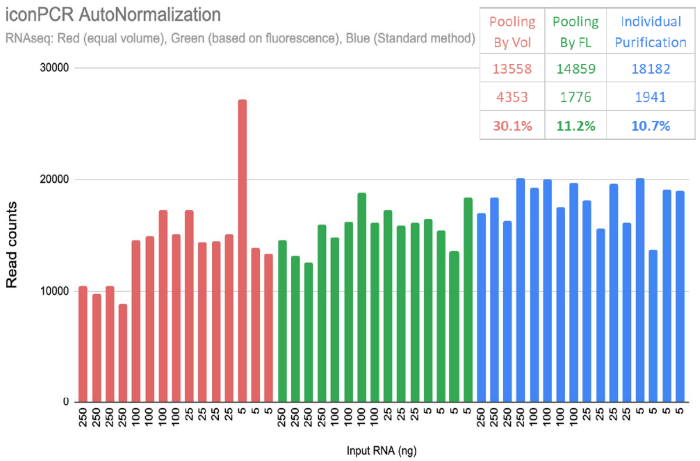
Image Credit: n6
Pooling by FL enables equal library balancing as a manual approach:
Adjusting the pooling volume according to the End-Point RFU values may reach an identical level to the laborious legacy approach. N6 produces an analysis tool to make direct recommendations for all approaches.
About n6
n6 proudly introduces iconPCR, a pioneering advancement in the genomics field with the world’s first real-time thermocycler with 96 individually controlled wells. This breakthrough technology promises to revolutionize DNA amplification and sequencing by offering unmatched simplicity and flexibility, setting a new standard for genomic research and diagnostics.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.