Austin W, Jouvenot Y, Patel P n6
When carrying out conventional PCR workflows users have to specify how many amplification cycles they need according to input quantity and assay type. This rigid structure is particularly difficult for formalin-fixed, paraffin-embedded (FFPE) samples, where nucleic acid degradation results in highly variable input quality and unpredictable amplification efficiency.
As a result, accurately quantifying these results is challenging, often requiring several thermocycler runs according to input amount and integrity, alongside labor-intensive pre- and post-PCR quality control steps. iconPCR, produced by n6, offers a fundamental change in PCR amplification with its AutoNorm technology.
By tracking fluorescence in real-time, iconPCR automatically concludes each reaction when a prespecified amplification threshold is attained. This per-sample, real-time control removes fixed-cycle setting requirements and regulates output across samples, notwithstanding input variability or degradation level.
iconPCR simplifies FFPE, and other difficult inputs, diminishing batch effects and improving both the consistency and quality of sequencing libraries, facilitating downstream analysis that is more efficient and precise in research and clinic alike.
Below, n6 presents two studies that showcase workflow and data quality enhancements for FFPE NGS preparations:
Study 1: Optimized whole genome sequencing of FFPE samples
Study 2: Effect of overamplification during RNA-seq of FFPE samples
iconPCR with AutoNorm overview
Engineering revolution
iconPCR
iconPCR is the world’s first thermocycler with individually controlled wells.

Image Credit: n6
With a fundamental change in hardware design, iconPCR is able to control the temperature and cycling of each well by itself.
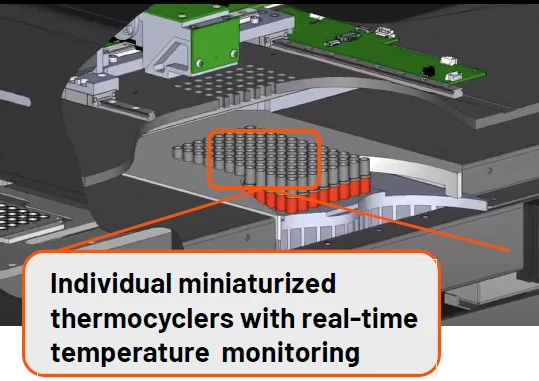
Image Credit: n6
Using AutoNorm for controlling amplification
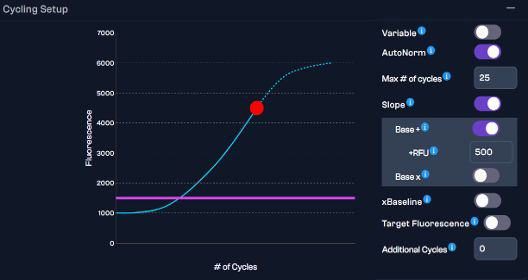
Image Credit: n6
Inside the iconPCR software, user-specified parameters can be used to guarantee target amplification without over-amplification or drop-outs from under-amplification.
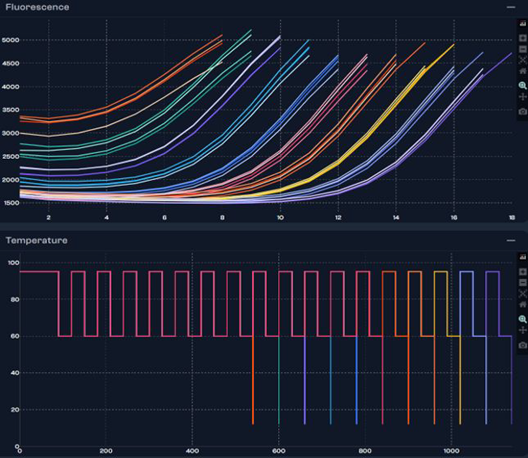
Image Credit: n6
The well stops cycling when the threshold is attained and transitions into a cold hold. The wells that remain continue cycling until they each reach their threshold.
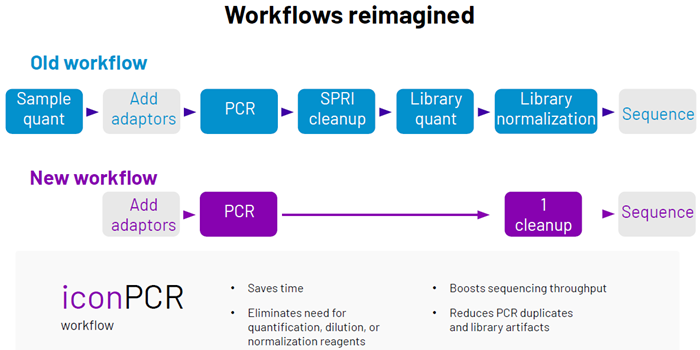
Image Credit: n6
Study 1: Optimized whole genome sequencing of FFPE samples
Details: Two FFPE-derived DNA samples (colon and liver) alongside a high-quality genomic DNA control (NA12878) were used to produce whole-genome sequencing (WGS) libraries at three input quantities: one ng, 10 ng, and 100 ng. Library construction was carried out using the manufacturer’s standard PCR protocol or iconPCR with AutoNorm.
After the PCR assays were carried out, the libraries underwent SPRI bead purification and quantification using Qubit fluorometry. Final libraries were sequenced using an Illumina NovaSeq X platform. Quality metrics, including library yield, coverage uniformity, and duplication rates, were measured to evaluate the effects of AutoNorm in comparison with standard fixed-cycle PCR.
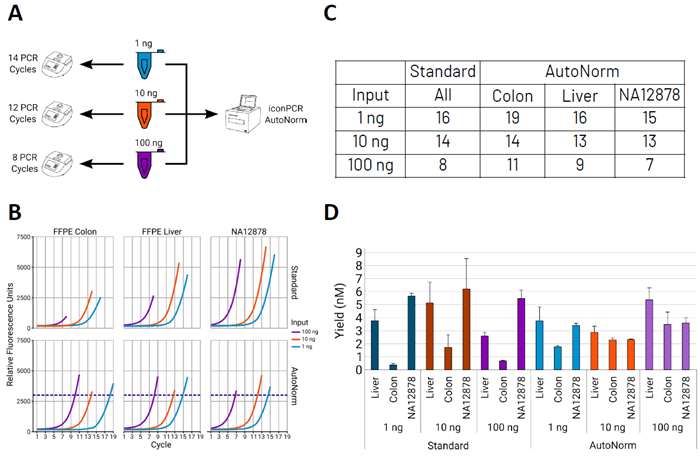
Figure 1. AutoNorm-enabled iconPCR accounts for sample diversity and streamlines complex workflows. Image Credit: n6
A) Whole-genome sequencing (WGS) libraries were generated using 1 ng, 10 ng, or 100 ng of input DNA. Traditional PCR workflows would require three separate instruments to optimize cycle numbers for each input, whereas iconPCR enables all libraries to be processed on a single instrument through AutoNorm. B) Representative amplification curves for each sample/input combination. Even with cycle adjustments for input amount, endpoint fluorescence values differ across inputs when using standard PCR.
Sample-specific variation is more pronounced than input variation. The colon FFPE sample exhibits the lowest amplification signal, followed by liver, with NA12878 showing the highest signal. C) Summary of PCR cycle requirements. NA12878 consistently required fewer cycles than recommended, while the colon sample needed additional cycles across inputs. The liver sample displayed mixed results, requiring extra cycles for 100 ng but fewer for 10 ng input. D) Final library yields.
Standard PCR conditions produced high variability across samples, with the colon sample generating low yields. In contrast, AutoNorm generated more uniform library concentrations. By dynamically applying additional cycles to the colon sample, AutoNorm increased yields and improved normalization prior to sequencing.
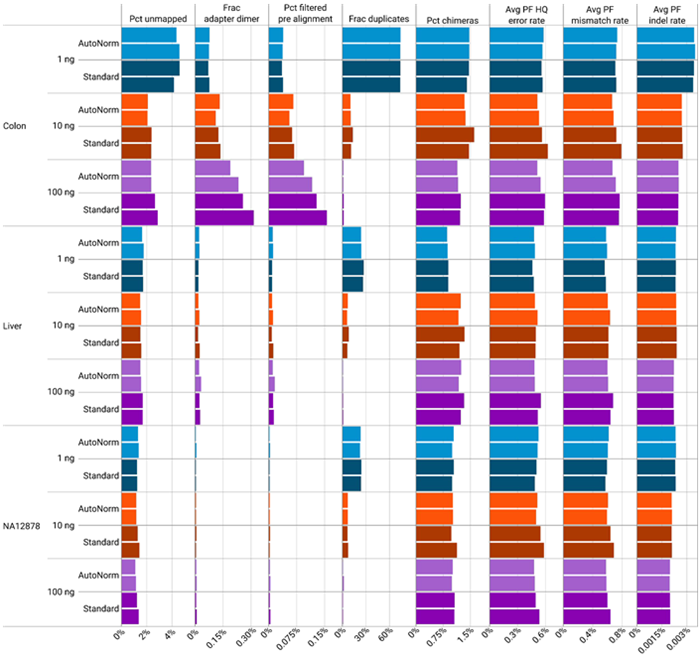
Figure 2. AutoNorm maintains sequencing quality metrics. Image Credit: n6
Quality metrics varied by both input amount and sample type. Libraries generated from one nanogram input exhibited the highest number of PCR duplicates and unmapped reads. Among sample types, the colon FFPE samples showed a higher percentage of unmapped reads and adapter dimers compared to liver or NA12878. Notably, AutoNorm did not negatively affect sequencing quality metrics, maintaining performance comparable to or better than standard PCR conditions.
Study 2: Effect of overamplification on FFPE RNA-seq data
Details: For Figure 1, RNA-seq libraries were prepared from 50 ng of RNA taken from a single FFPE sample using a spectrum of PCR cycle numbers. Libraries were sequenced and then aligned, and quality metrics (e.g., read mapping rates, duplication levels) alongside gene counts were calculated.
For Figure 2, RNA-seq libraries were readied using 1 ng, 10 ng, and 100 ng of RNA from four different FFPE samples. Libraries were amplified via the manufacturer’s recommended fixed PCR cycles (standard) or iconPCR with AutoNorm. After sequencing and alignment, identical quality metrics and gene count analyses to those outlined for Figure 1 were carried out.
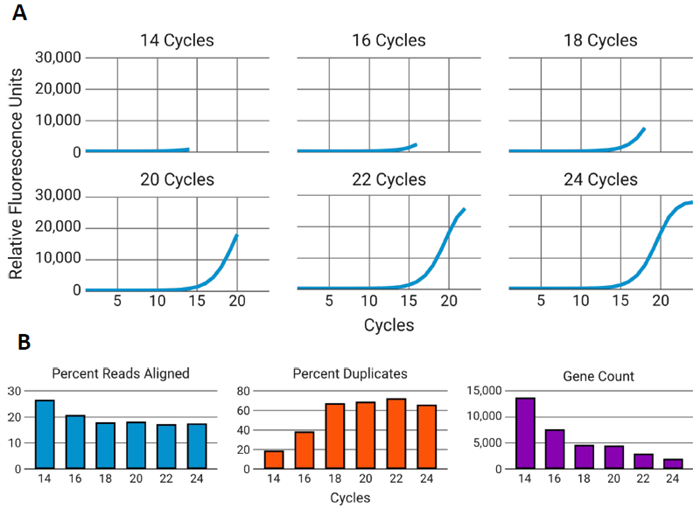
Figure 3. Overamplification of RNAseq libraries reduces data quality. Image Credit: n6
A) RNA-seq libraries were generated from 50 ng of RNA from a single FFPE sample, with amplification ranging from 14 to 24 PCR cycles in 2-cycle increments. Each panel represents the point along the amplification curve where cycling was terminated. B) Libraries were sequenced, subsampled to 1 million reads, and analyzed. As the number of PCR cycles increased, there was a decrease in the percentage of aligned reads (left), an increase in PCR duplicates (center), and a reduction in detected gene counts (right).
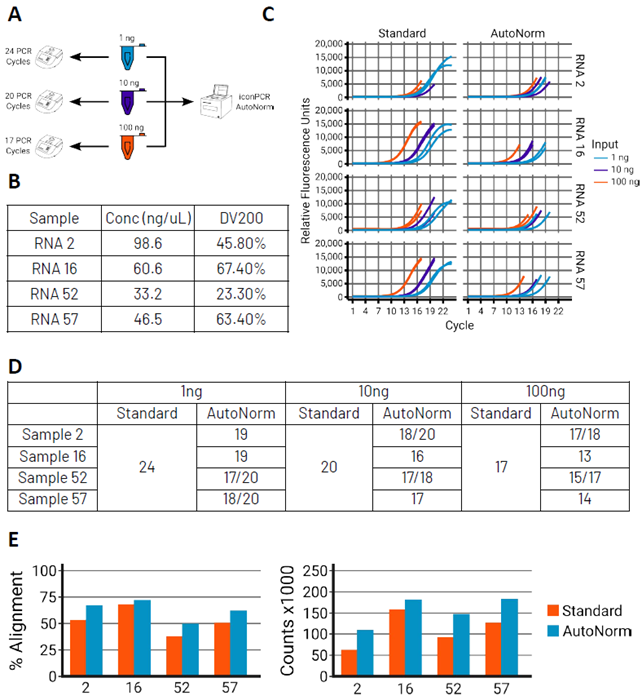
Figure 4. AutoNorm preserves data integrity by preventing overamplification. Image Credit: n6
A) Standard PCR workflows require dividing samples of different input amounts across multiple thermocyclers, each programmed with a different number of PCR cycles. In contrast, iconPCR with AutoNorm enables all libraries, regardless of input amount, to be amplified on a single instrument. B) Table summarizing the FFPE samples used in this study, including initial RNA concentrations and DV200 values as a measure of RNA integrity. C) Real-time PCR amplification curves using standard PCR cycling (left) versus iconPCR with AutoNorm (right).
Standard cycling results in variable endpoint fluorescence due to input and sample quality differences. With AutoNorm, each sample terminates at the same amplification threshold, independent of input or degradation level. D) Number of PCR cycles required for each sample. E) AutoNorm improves data quality. Libraries from 1 ng input were sequenced and subsampled to 1M reads. Graphs show the percentage of reads aligning to the reference genome and the number of deduplicated gene counts, averaged across two technical replicates.
About n6
n6 proudly introduces iconPCR, a pioneering advancement in the genomics field with the world’s first real-time thermocycler with 96 individually controlled wells. This breakthrough technology promises to revolutionize DNA amplification and sequencing by offering unmatched simplicity and flexibility, setting a new standard for genomic research and diagnostics.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net, which is to educate and inform site visitors interested in medical research, science, medical devices, and treatments.