Li, P.1, Austin W.2, Patel P.2, Jouvenot Y.2
1: Rush University Genomics & Microbiome Core Facility (GMCF), Chicago, IL 60612
2: n6, Pleasanton, CA 94566
Overview
With standard PCR systems, users are required to set a predefined quantity of cycles according to assay and sample-specific factors. Single-cell sequencing tests demand knowledge of the number of cells being recorded alongside RNA content within cells. Different cell numbers and RNA content need different numbers of PCR cycles, necessitating several thermocycler runs, making workflows more complex and increasing the number of potential sources of error.
iconPCR, built by n6, constitutes a significant shift in PCR technology. Its central innovation arises from AutoNorm™, wherein every PCR reaction is overseen and concluded in real-time according to fluorescence thresholds.
This removes the guesswork inherent to fixed-cycle PCR and ensures that each library is amplified in the best way. iconPCR enables novel single-cell workflows, making it possible to run samples of different types and inputs at the same time, which increases efficiency and protects against incorrect cycling conditions.

Figure 1. iconPCR, the world’s first real-time thermocycler with 96 individually controlled wells. Image Credit: n6
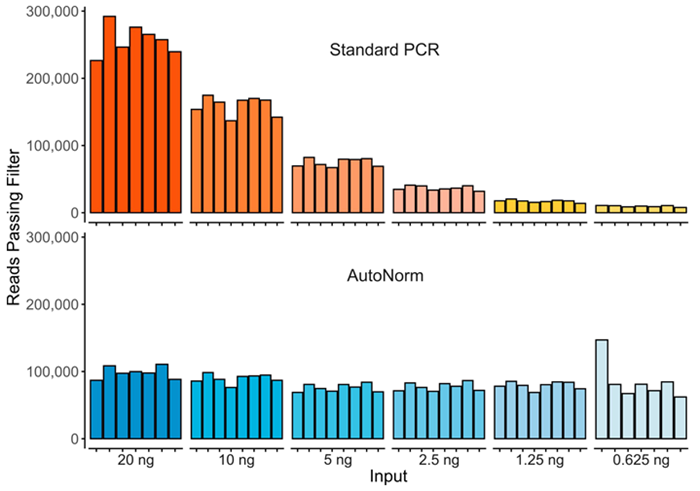
Figure 2. Here we show the significant yield variance when using a single PCR instrument with a fixed number of PCR cycles compared to iconPCR with AutoNorm where each sample is amplified to similar levels. Image Credit: n6
To assess performance, n6 prepared a series of 10x Genomics GEM-X Universal 3’ Gene Expression v4 libraries with a broad spectrum of cell inputs. After cell capture and preamplification, the cDNA and libraries were prepared via the standard protocol or the iconPCR tool via AutoNorm. iconPCR simplified workflow while sustaining the quality of single-cell gene expression data.
Experimental design
A single culture of cells was recorded using two replicate GEM-X chips. For each chip, two wells of 500, 3000, 6000, and 10,000 cells were recorded, providing 16 samples altogether. Half of the samples underwent the standard workflow, while the other half underwent an adjusted workflow with iconPCR.
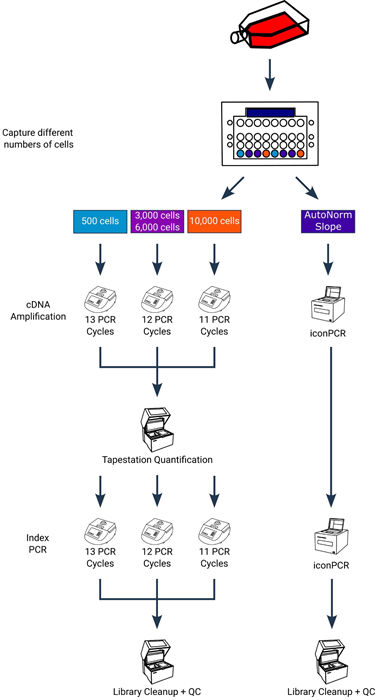
Figure 3. Experimental design showing critical workflow differences. Following cell capture and preamplification, the cDNA going through the standard workflow (left side) was run across 3 different PCR runs, in accordance with the standard PCR cycling conditions. Following cDNA amplification each sample was quantified in order to determine the appropriate number of cycles for index PCR and amplified accordingly. For cDNA amplification on iconPCR (right side), all 8 samples were run together on a single instrument using the slope method of AutoNorm. Following cDNA amplification, samples were again run together using the slope method of AutoNorm for index PCR. Following cleanup and QC all libraries were pooled for sequencing. Image Credit: n6
Reduced workflow complexity
Standard PCR needed three separate thermocycler runs for cDNA amplification and index PCR. In contrast, all samples, regardless of input amount, were run on the iconPCR at the same time in both steps.
With AutoNorm, cycle numbers were calculated dynamically, and each well stopped when the fluorescence reached the linear amplification curve’s maximum slope.
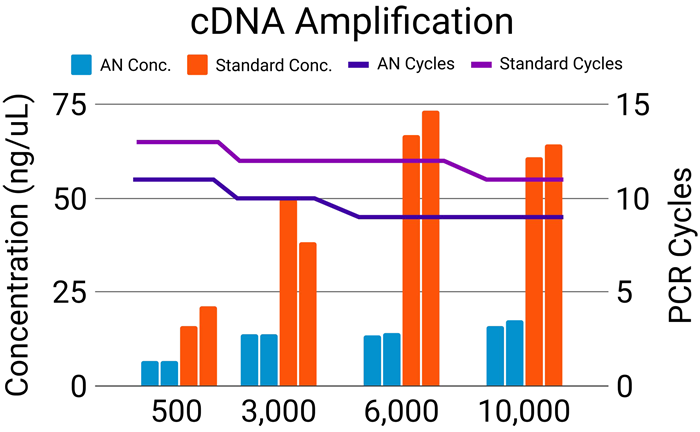
Figure 4. AutoNorm (AN) allows for the efficient processing of samples across a range of inputs. AutoNorm resulted in a 2-3 cycle decrease as compared to the standard PCR cycling recommendations (navy vs purple lines). A wide range of concentrations was seen across the various inputs when using standard PCR conditions (orange bars). In comparison, iconPCR with AutoNorm provided a more even final concentration (light blue bars). It’s important to note that although the concentration post cDNA amplification was determined for all samples, this is not a requirement to proceed with index PCR when using AutoNorm. Image Credit: n6
Table 1. AutoNorm simplifies index PCR. Due to differences in concentration following cDNA amplification, the standard protocol required the use of 3 separate PCR runs. Conversely, iconPCR samples were able to be run on a single instrument. Use of AutoNorm reduced the number of cycles by 2-6 compared to the recommendations in the standard protocol. Source: n6
| Chip |
Input (cells) |
Input for Index PCR (ng) |
Index PCR cycles |
Standard recommended cycle number |
Cycle Difference |
| Chip 1 |
500 |
65.5 |
10 |
12-14 |
2-4 |
| 3000 |
136.5 |
9 |
12-14 |
3-5 |
| 6000 |
135 |
9 |
12-14 |
3-5 |
| 10000 |
159 |
8 |
12-14 |
4-6 |
| Chip 2 |
500 |
66.5 |
9 |
12-14 |
3-5 |
| 3000 |
137.5 |
9 |
12-14 |
3-5 |
| 6000 |
141 |
8 |
12-14 |
4-6 |
| 10000 |
176.5 |
8 |
12-14 |
4-6 |
Table 2. Summary of PCR Cycling. iconPCR with AutoNorm (AN) resulted in an overall decrease of 5-6 cycles of PCR as compared to the standard PCR protocol. In total, the standard protocol required 6 separate thermocycler runs, while use of iconPCR reduced that to 2 total runs, showcasing iconPCR’s ability to simplify complex workflows. Source: n6
| Chip |
Method |
Input |
cDNA Amp Cycles |
Index PCR Cycles |
Total Cycles |
| Chip 1 |
AutoNorm |
500 |
11 |
10 |
21 |
| AutoNorm |
3000 |
10 |
9 |
19 |
| AutoNorm |
6000 |
9 |
9 |
18 |
| AutoNorm |
10000 |
9 |
8 |
17 |
| Standard |
500 |
13 |
13 |
26 |
| Standard |
3000 |
12 |
12 |
24 |
| Standard |
6000 |
12 |
11 |
23 |
| Standard |
10000 |
11 |
11 |
22 |
| Chip 2 |
AutoNorm |
500 |
11 |
9 |
20 |
| AutoNorm |
3000 |
10 |
9 |
19 |
| AutoNorm |
6000 |
9 |
8 |
17 |
| Auto Norm |
10000 |
9 |
8 |
17 |
| Standard |
500 |
13 |
13 |
26 |
| Standard |
3000 |
12 |
12 |
24 |
| Standard |
6000 |
12 |
11 |
23 |
| Standard |
10000 |
11 |
11 |
22 |
No loss of data quality
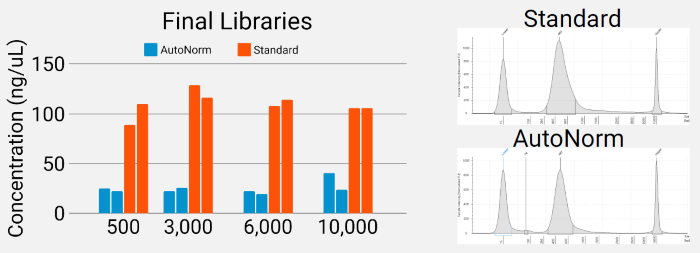
Figure 5. Final library quality control. AutoNormalized libraries provided sufficient quantities of final library for sequencing (left) and maintain the same quality as determined by Tapestation (right). Image Credit: n6
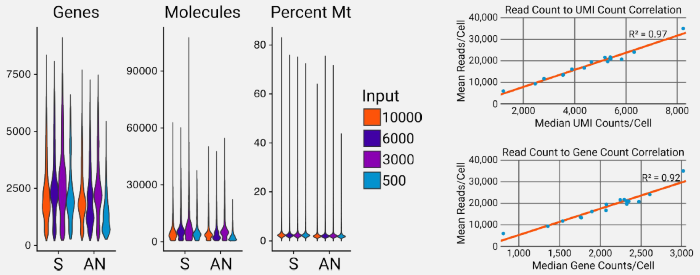
Figure 6. AutoNorm samples (AN) maintain the same quality as libraries prepared using the standard procedure (S). AutoNormalized libraries and libraries prepared using the standard protocol showed similar results in the number of genes and unique molecules (UMIs) detected, and did not change critical quality metrics such as the percent of mitochondrial reads (% mito). All samples showed a strong correlation between the mean reads per cell and both the median UMI counts per cell and median genes per cell, indicating that any difference in gene and UMI counts is due to differences in sequence depth between samples. Image Credit: n6
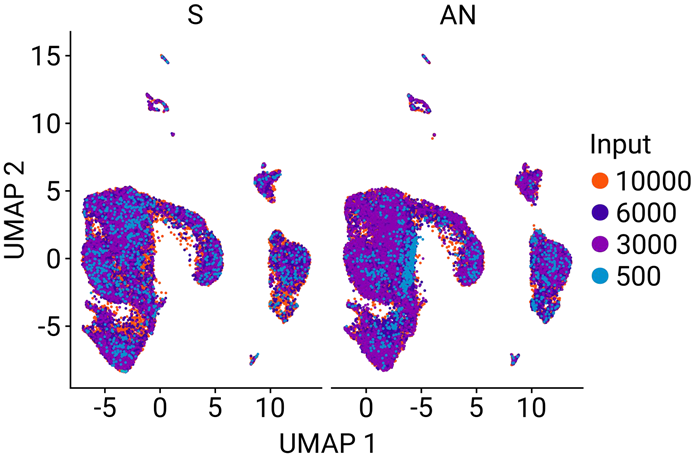
Figure 7. Clustering of all samples reveals common gene signatures. Clustering of all 16 samples revealed no differences between the standard PCR method (S) and .AutoNorm (AN), showcasing that the change in workflow does not negatively affect data quality. Image Credit: n6
Advantages of iconPCR. Source: n6
| Conventional PCR |
iconPCR (with AutoNorm) |
| Fixed cycle count (e.g., 30 cycles) |
Real-time fluorescence monitoring |
| One-size-fits-all amplification |
Per-well cycle control based on signal, not guesswork |
| Under/over-amplification common |
Optimal amplification per sample |
| High chimera rates |
Reduced chimera formation |
| Variable library quality |
Uniform library quality across all wells |
| Manual quant and normalization required |
Automated normalization (no post-PCR quant) |
| More hands-on time |
40–60 % reduction in hands-on time |
| Increased reagent waste |
Lower reagent waste, fewer failed libraries |
| Extra QC and rerun costs |
Faster turnaround, minimal QC/rescue steps |
Conclusion
iconPCR transforms single-cell library preparation by eradicating the limitations of conventional PCR workflows. With its unique per-well AutoNorm mechanism, iconPCR produces optimized PCR cycling regardless of cell input.
Samples of different cell numbers no longer need to be divided among thermocyclers, and experiments are no longer at risk when the number of cells recorded differ from expectation.
Use of iconPCR minimizes complexity, enabling a higher throughput of samples for processing with a smaller chance of error and without loss of data quality.
About n6
n6 proudly introduces iconPCR, a pioneering advancement in the genomics field with the world’s first real-time thermocycler with 96 individually controlled wells. This breakthrough technology promises to revolutionize DNA amplification and sequencing by offering unmatched simplicity and flexibility, setting a new standard for genomic research and diagnostics.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.