Overview
Standard PCR systems require users to set a predefined number of cycles relative to the assay and the input amount. This requires quantifying input material and splitting samples of varying inputs over different PCR runs.
Despite this, outputs vary widely and need to be purified, which is a time-consuming and costly process, as well as quantification and normalization techniques to reach proper pool balancing before sequencing.
n6’s iconPCR™ represents a dramatic shift in PCR technology. This shift is predominantly centred around AutoNormalization (AutoNorm™), in which each PCR reaction is tracked and concluded in real time according to fluorescence thresholds.
This eliminates the guesswork typical of fixed-cycle PCR and ensures that each library is amplified in the best way and to equivalent amounts. AutoNorm enables the pooling of libraries to take place before purification, simplifying the workflow and reducing associated costs
To assess performance, n6 prepared whole genome sequencing libraries from a single sample over a broad spectrum of input quantities. After amplification with AutoNorm, an equal volume of each sample was pooled, purified, and then sequenced to calculate read balancing. Libraries pooled according to AutoNorm demonstrated a similar read distribution to a pool produced via the standard process, but with reduced hands-on time and expense.

Figure 1. iconPCR, the world’s first real-time thermocycler with 96 individually controlled wells. Image Credit: n6
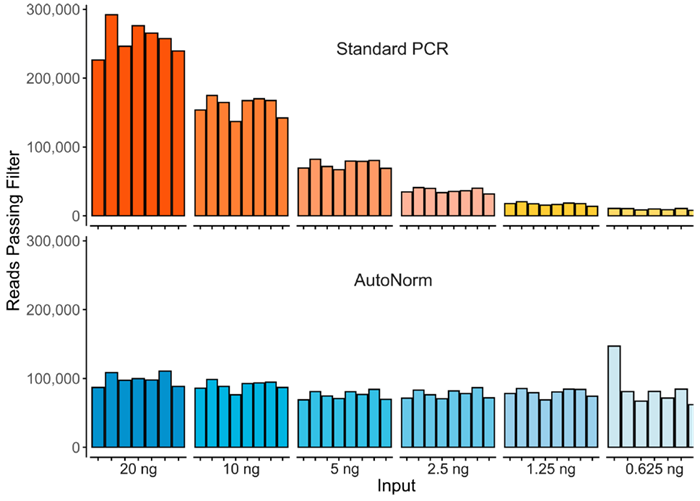
Figure 2. Here we show the significant yield variance when using a single PCR instrument with a fixed number of PCR cycles compared to iconPCR with AutoNorm, where each sample is amplified to similar levels. Image Credit: n6
Experimental design
A dilution series spanning from ten to 120 ng in ten nanogram increments was generated on a 96-well plate. Libraries were prepared using the standard PCR protocol, and separately on iconPCR with AutoNorm to compare differences.
Optimized workflow
Standard PCR necessitated three different thermocycler runs of six, seven, and eight cycles, linked to inputs of ten to 20 ng, 30 to 70 ng, and 80 to 120 ng. With iconPCR assays, all of the samples, regardless of input quantity, were run on the iconPCR at the same time. With AutoNorm, cycle numbers could be calculated dynamically, with each well stopping when fluorescence reached its target value of 6000 relative fluorescence units (RFU).
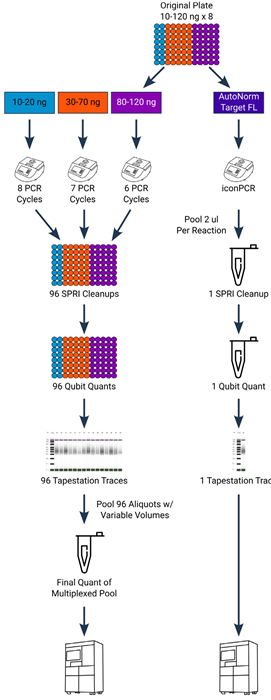
Figure 3. Experimental design showing simplified workflow for iconPCR. Standard PCR (left side) required samples of varying inputs to be split across multiple different PCR runs. Following amplification, each sample must be individually purified, quantified, and normalized, and then pooled prior to sequencing. Conversely, with iconPCR, all samples were processed simultaneously. Following amplification, two μL of each sample were pooled into a single tube, purified, quantified, and then loaded onto the sequencer. Image Credit: n6
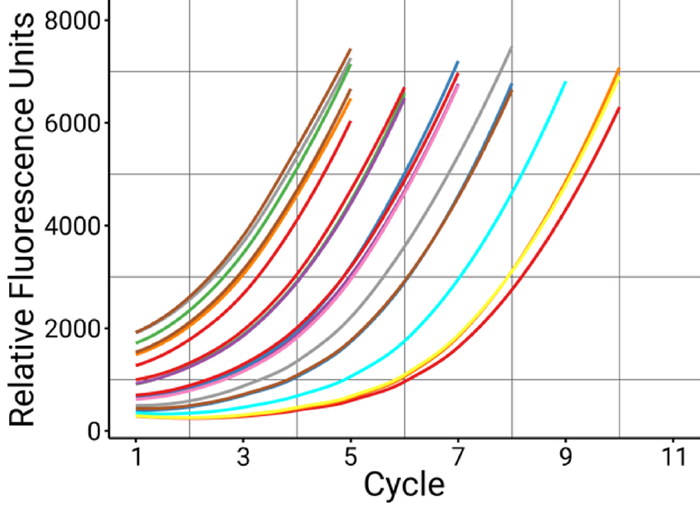
Figure 4. Proper amplification of samples in a single run with AutoNorm. All samples, regardless of input amount, showed a consistent amplification stopping point, showcasing the ability of iconPCR to individually control cycling conditions on independent wells. Image Credit: n6
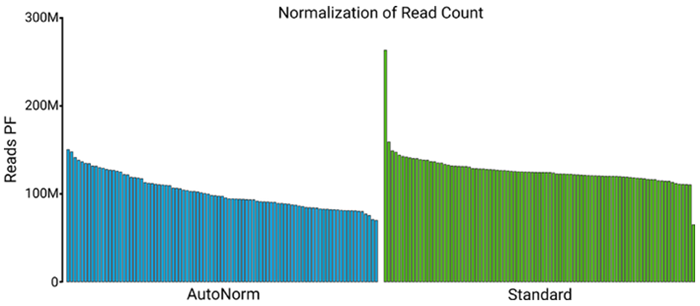
Figure 5. Number of reads passing filter. Ranked bar plots show the number of reads passing the filter following the sequencing of the AutoNorm and Standard workflow library pools. A similar read distribution was observed between the methods. Image Credit: n6
Sample recovery
Variance in sample quality, pipetting errors, and mathematical errors in dilution calculations can result in library preparation failures. In this study, seven samples failed library generation in the standard PCR protocol. Six were rescued via iconPCR.
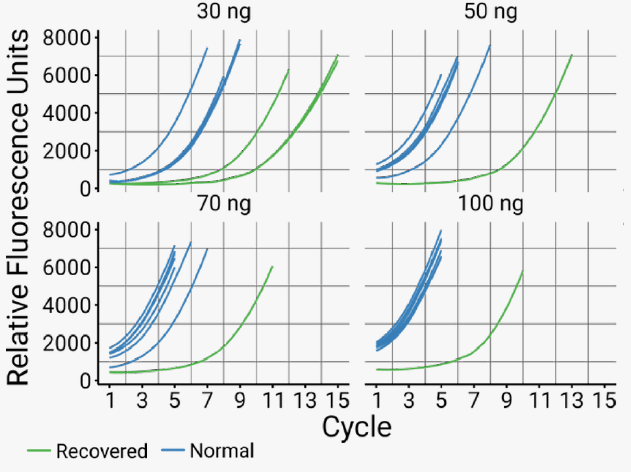
Figure 6. Amplification curves of recovered samples. The PCR amplification curves of the six rescued samples (green) show that each needed several additional cycles to reach similar levels of amplification as samples that successfully generated libraries in both PCR approaches (blue). Image Credit: n6
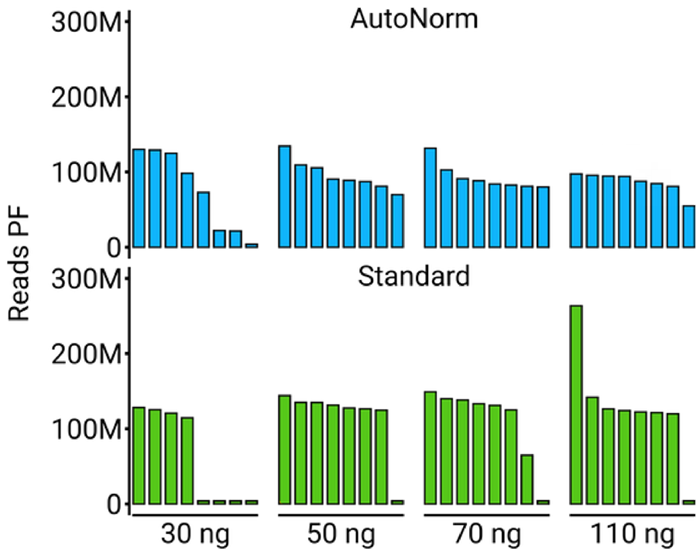
Figure 7. Number of reads passing the filter of recovered samples. Barplots show the number of reads passing the filter for the four concentrations containing failed libraries. AutoNorm rescued six of the seven, with only one library at the 30 ng input amount also failing. Image Credit: n6
Source: n6
| Conventional PCR |
iconPCR (with AutoNorm) |
| Fixed cycle count (e.g., 30 cycles) |
Real-time fluorescence monitoring |
| One-size-fits-all amplification |
Per-well cycle control based on signal, not guesswork |
| Under/over-amplification common |
Optimal amplification per sample |
| High chimera rates |
Reduced chimera formation |
| Variable library quality |
Uniform library quality across all wells |
| Manual quant and normalization required |
Automated normalization (no post-PCR quant) |
| More hands-on time |
40-60 % reduction in hands-on time |
| Increased reagent waste |
Lower reagent waste, fewer failed libraries |
| Extra QC and rerun costs |
Faster turnaround, minimal QC/rescue steps |
Conclusion
iconPCR overcomes the limitations of conventional PCR workflows with its per-well AutoNorm™ system, reducing hands-on time and reagent costs while maintaining balanced reads for maximum flow cell use.
It maximizes lab operations by restricting the number of failed libraries and eliminating costly and time-consuming sample requests.
About n6
n6 proudly introduces iconPCR, a pioneering advancement in the genomics field with the world’s first real-time thermocycler with 96 individually controlled wells. This breakthrough technology promises to revolutionize DNA amplification and sequencing by offering unmatched simplicity and flexibility, setting a new standard for genomic research and diagnostics.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net which is to educate and inform site visitors interested in medical research, science, medical devices and treatments.