Optimizing PCR assays for assay development (AD) workflows typically involves a range of interdependent factors. These include thermocycling conditions, enzyme levels, buffer chemistry, and oligonucleotide concentrations.
Most traditional thermocyclers limit flexibility because each well on a given plate must have identical cycling conditions. As a result, researchers have to split tests across runs, inflating workloads, consumable use, and extending turnaround times.
Design of Experiments (DOE) is a powerful framework for dealing with complex parameter spaces in AD workflows. A study by Whitcomb & Kraber (Stat-Ease) showed optimization over nine factors, including three thermocycler settings and six reagent parameters. On legacy thermocyclers, such research requires split-plot designs in which thermocycler factors are fixed over whole plates.1
The iconPCR platform gets rid of this constraint. With 96 independently-controlled wells, linear gradients exceeding 80 °C, and integrated AutoNorm™ real-time normalization, iconPCR allows diverse thermocycler conditions to be optimized inside just one plate. The following discusses how to carry out Whitcomb & Kraber’s DOE on iconPCR:
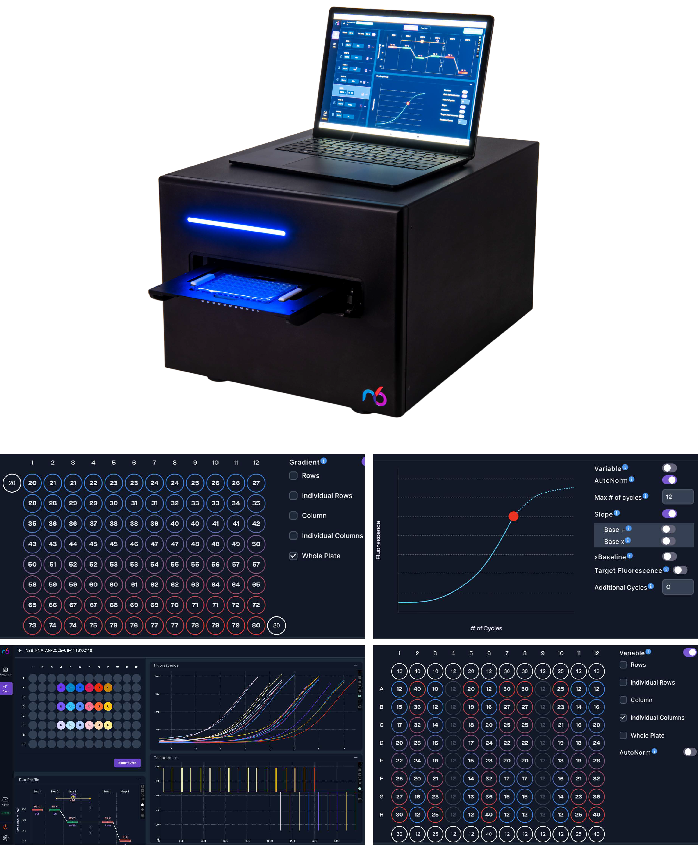
Figure 1. The iconPCR instrument and software interface. A compact benchtop system, iconPCR combines thermocycling and real-time analysis with: • 96 independent wells for fully randomized DOE designs. • Linear gradients enabling broad temperature sweeps. • AutoNorm for automated per-well endpoint normalization. • Intuitive visualization tools for amplification curves, temperature profiles, and reagent conditions. Image Credit: n6
DOE workload comparison
- In the Whitcomb & Kraber DOE design, six reagent parameters can be tested in a single PCR plate: forward primer concentration, reverse primer concentration, DNA probe concentration, MgCl2 concentration, Tween concentration, and polymerase concentration.
- Legacy thermocyclers: Although some models include options for temperature gradients, they lack complete per-well control. As 32 wells per plate are consumed by reagent combinations, it is not possible to apply gradient features at the same time. As a result, each thermocycler condition (annealing temperature, denature time and temperature) needs to be run on different plates. (Figure 2A).
- iconPCR: Independent well control enables various annealing and denature temperatures to run on a single plate (Figure 2B), reducing the number of plates needed by half. Unlike traditional systems, edge effects are eliminated with iconPCR as each well is independently tracked and accurately maintained at a predetermined temperature, making it possible to use the whole plate.
Scalability example
Adding just one annealing temperature to the DOE heightens the number of plates on legacy thermocyclers by 50 % (Figure 2C). With iconPCR, the same additional temperature may be integrated into unused wells without adding more plates (Figure 2D).
Scaling the quantity of factors further demonstrates the effects of iconPCR.
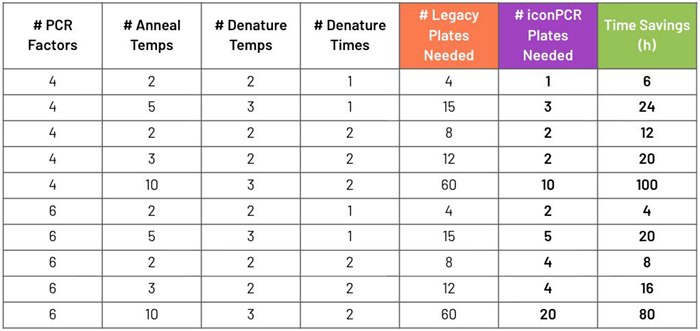
For instance, a design testing six PCR factors alongside five annealing and three denature conditions would demand 15 plates on a legacy system, and just five plates on iconPCR, which could save 20 hours (h) in total.
At most, designs with 10 annealing temperatures and diverse denaturation settings may require 60 plates on legacy thermocyclers, yet just 10-20 plates on iconPCR. In such cases, the time savings can extend beyond 80-100 h, on top of the significant decrease in consumable use (Table 1).
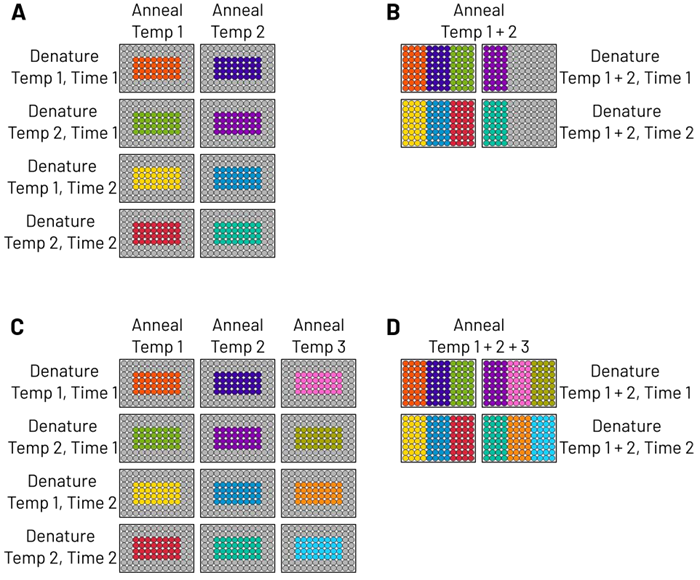
Figure 2. Example of DOE plate setup A) Legacy thermocycler conditions are locked plate-wide, requiring eight separate PCR runs. B) iconPCR allows multiple annealing and denature conditions in a single plate, reducing runs by 50 %. C) Adding an extra annealing temperature to legacy systems increases plates by 50 %, totaling 12 runs. D) Addition of another annealing temperature can be incorporated into existing plates with no increase in runs on iconPCR. Image Credit: n6
Table 1. Plates required for DOE on legacy thermocyclers vs. iconPCR. Time savings calculated at two hours per plate. Across these representative designs, iconPCR reduces plate usage by 50–80 %, corresponding to up to 100 h of time saved depending on study size. Source: n6
Concluding remarks
iconPCR is transforming PCR assay development. DOE research that at one point required dozens of plates and long hours at the thermocycler can now be undertaken with a small number of plates, less hands-on time, and better data, all while reducing the use of consumables.
By removing split-plot obstacles and constructing per-well normalization, iconPCR makes full-factorial optimization both practical and scalable. For assay developers, this leads to faster iteration, more intelligent reagent use, and better-quality data at every step.
References and further reading
- Whitcomb, P. and Kraber, S. PCR Process Optimized via Split-Plot DOE. (online) Available at: https://cdnm.statease.com/pubs/pcr_via_split-plot.pdf.
About n6
n6 proudly introduces iconPCR, a pioneering advancement in the genomics field with the world’s first real-time thermocycler with 96 individually controlled wells. This breakthrough technology promises to revolutionize DNA amplification and sequencing by offering unmatched simplicity and flexibility, setting a new standard for genomic research and diagnostics.
Sponsored Content Policy: News-Medical.net publishes articles and related content that may be derived from sources where we have existing commercial relationships, provided such content adds value to the core editorial ethos of News-Medical.Net, which is to educate and inform site visitors interested in medical research, science, medical devices, and treatments.