Researchers in Canada have discovered two novel variants in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) gene sequences collected from mink in the United States.
The SARS-CoV-2 virus is the agent responsible for the coronavirus disease 2019 (COVID-19) pandemic that has now claimed the lives of more than 2.78 million people globally.
The team identified a two-mutation (N501T-G142D) variant and a three-mutation (N501T-G142D-F486L) variant in the SARS-CoV-2 spike protein – the main structure the virus uses to bind to and infect cells.
The receptor-binding domain (RBD) of the spike attaches to the host cell receptor angiotensin-converting enzyme 2 (ACE2) to enable viral fusion and cell entry.
The two variants were not found in mink-derived SARS-CoV-2 gene sequences collected in any other countries.
Hugh Cai from the University of Guelph in Ontario and Allison Cai from the University of British Colombia say the findings suggest that the variants evolved during human infection before they spread to mink populations.
The team says it is vital to monitor the emerging new variants to determine their impact on human and animal health.
A pre-print version of the research paper is available on the medRxiv* server, while the article undergoes peer review.
.jpg)
Study: SARS-CoV-2 spike protein gene variants with N501T and G142D mutation dominated infections in minks in the US. Image Credit: Gallinago_media / Shutterstock

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The virus has rapidly evolved mutations in the spike protein
Since the first SARS-CoV-2 sequence was isolated from samples collected in Wuhan, China, in December 2019, the virus has rapidly evolved spike protein mutations that have increased its transmissibility and virulence.
As of 17th March 2021, 22% of SARS-CoV-2 genomes within the GISAID (Global Initiative on Sharing All Influenza Data) database contained the N501Y mutation. This variant has been identified in the spike protein of the more transmissible and potentially more virulent lineages that recently emerged in the UK (B.1.1.7), South Africa (B.1.351), and Brazil (B.1.525).
In Denmark, large numbers of mink were also found to be infected with SARS-CoV-2 containing the spike mutation Y453F, which triggered concerns that this variant could be spread from animals to humans.
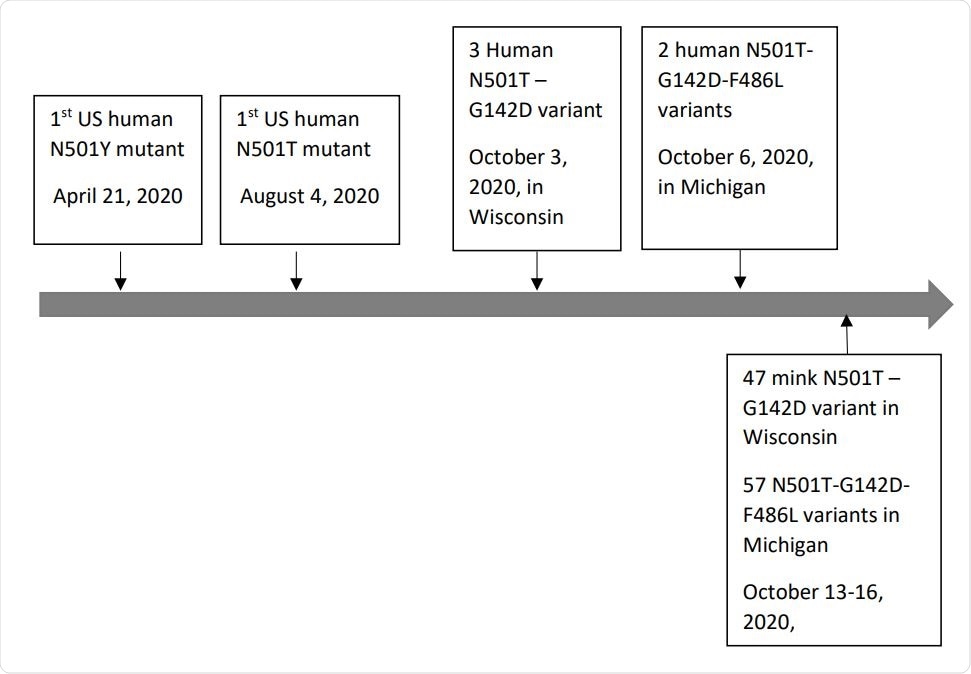
Emerging timeline of human-derived (above the timeline) and mink-derived (below the timeline) SARS-CoV-2 spike protein N501T-G142D and N501T-G142D-F486L variants in the US. Timeline scale is not in proportion.
What did the researchers do?
The researchers set out to determine the genetic characteristics of SARS-CoV-2 spike protein sequences collected from mink in the United States and Canada.
The team analyzed all animal-derived SARS-CoV-2 sequences (977), all sequences collected in Canada (19,529), and all sequences collected in the United States (173,277) that were deposited in GISAID between December 2019 and 12th March 2021.
The SARS-CoV-2 isolate Wuhan-Hu-1 collected on 19th December 2019 was used as a reference for mutation analysis.
The “BLAST” function of the bioinformatics software “Geneious” was used to determine the incidence of any novel mutations locally and globally.
What did the analysis reveal?
Neither the N501Y mutation shared by the UK, South African, and Brazil variants nor the Y453F mutation identified among mink in Denmark was present in any of the US and Canadian mink-derived SARS-CoV-2 sequences.
However, the team identified two dominant novel variants in spike protein sequences collected from mink in the US – the double mutation N501T-G142D and the triple mutation N501T-G142D-F486L.
The variants were not found in sequences collected from mink in Canada or any other country.
To determine the origin of the novel variants, the researchers analyzed all human-derived SARS-CoV-2 sequences collected from the US and Canada.
This revealed that the N501T mutation (without G142D or F486L) was present in US human-derived sequences collected as early as August 2020 – two months prior to when the mink-derived sequences were collected (October 2020).
The team also says both the N501T-G142D and N501T-G142D-F486L variants were identified in the human-derived sequences before they were identified in the mink-derived sequences.
What are the implications of the study?
“The results of this study indicate that the novel variants may have evolved during human infection and then been transmitted to mink populations in the US,” writes the team.
The researchers say the occurrence of these new variants in the mink population warrants further study to establish how they may have affected spike’s interaction with ACE2 and, therefore, viral transmissibility, virulence, and immunogenicity in humans and mink.
“It is important to monitor the emerging new variants and determine their impact on human and animal health,” they conclude.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources