The B.1.1.7 variant was first detected in the United Kingdom last fall and has since spread globally. Now new reports are surfacing of the B.1.1.7 variant harboring the E484K mutation that is notorious for escaping neutralizing antibodies — including neutralizing activity from vaccine-induced immunity.
Their results also showed seven distinct groups with the E484K mutation on the spike protein, which raises the chances for seven ways for the B.1.1.7 lineage to acquire the mutation.
The research team writes:
“Phylogenetic analysis suggested the presence of at least 3 distinct clades of B.1.1.7+E484K circulating in the U.S., with the Pennsylvanian isolates belonging to two distinct clades. Increased genomic surveillance will be crucial for detection of emerging variants of concern that can escape natural and vaccine-induced immunity.”
The study “Comparative Analysis of Emerging B.1.1.7+E484K SARS-CoV-2 isolates from Pennsylvania” is available as a preprint on the bioRxiv* server, while the article undergoes peer review.
Rapid increase in B.1.1.7 in the Pennsylvania area
The researchers performed genomic sequencing on randomly selected isolates from nasopharyngeal swabs collected since January 2021. Of those, they sequenced 114 genomes. In March, the B.1.1.7, B.1.429 variant in California, the B.1.526 variant in New York, and R.1 variant made up 69% of the genome.
The B.1.1.7 variant jumped from 2% detected in February to 42% in March. One B.1.1.7 strain had the E484K mutation on the spike protein.
They used the public genomic database to compare their findings with other coverage of B.1.1.7 with the E484K mutation. The variant and mutation have also been found worldwide, with 253 genomes identified in England and 14 other countries, including the United States.
Prevalence of B.1.1.7 + E484K in the United States
To determine when the B.1.1.7 and E484K mutation spread in the United States, they mapped cases of it from December 2020 to March 2021. The first 60 cases detected were from February 6, 2021, in Oregon. Soon after, reports surfaced in 15 other states.
The majority of cases (48%) with B.1.1.7 and the E484K mutation came from Florida and New York. About 28% came from New Jersey, California, and Pennsylvania.
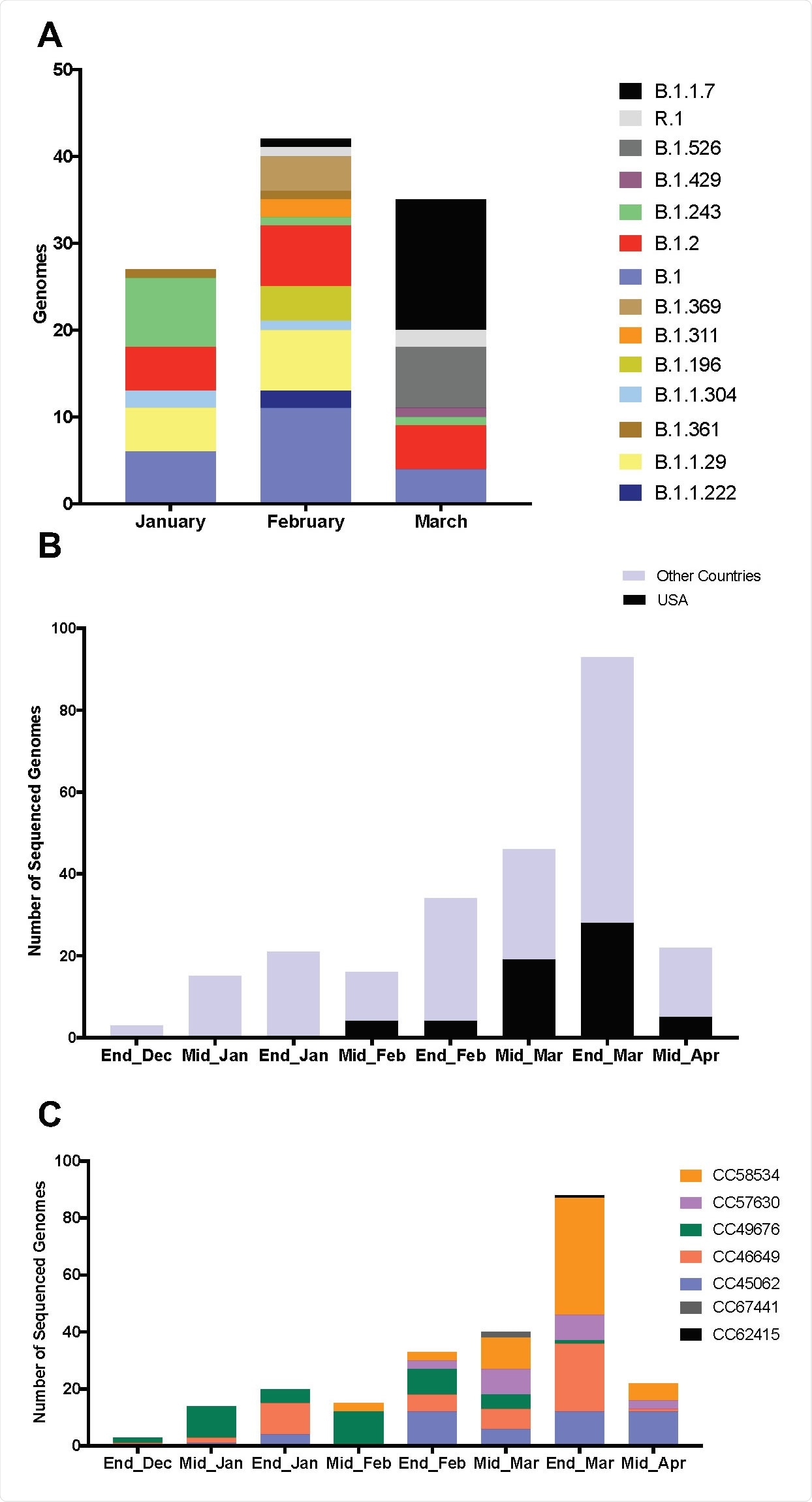
Diversity of SARS-CoV-2 in Philadelphia and global diversity of sequenced B.1.1.7+E484K genomes. A. Stacked bar plot showing the diversity of random genomes sequenced by our laboratory at Children’s Hospital of Philadelphia during January, February and March 2021. Ten lineages that were represented by only one genome (B.1.1, B.1.1.106, B.1.1.129, B.1.1.197, B.1.1.281, B.1.1.296, B.1.119, B.1.234, B.1.350, B.1.409) were excluded from the plot. One isolate that is B.1.526.1 was counted with the parent B.1.526 for easier visualization. B. Bar plot showing number of GISAID genomes (n=250) that are 20I/501Y.V1 and have the E484K spike mutation over time in the US and globally. C. Diversity of 236 isolates according to GNUVID. Bar plot showing relative abundance of circulating clonal complexes (CC) for the 236 B.1.1.7+E484K isolates (typed by GNUVID). The bar plot shows that the isolates belong to 7 different CCs. Isolate EPI_ISL_1385215 was not assigned to any of the 7 CCs (CC255). Fourteen isolates were excluded from the plot as they had > 5% nucleotides designated “N” in the sequence.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Similar to what was observed in Pennsylvania, the number of reports of the B.1.1.7 + E484K mutations rose from February to March. The number of isolates detected was six times higher than the previous month.
“This increase raises the concern that more B.1.1.7+E484K sequences may be emerging even as herd immunity increases by 98 natural immunity and vaccines,” suggests the research team.
B.1.1.7 lineages may have acquired the E484K mutation independently
Further analysis suggests the E484K mutation developed independently of other nearby strains with the mutation. It shows at least three B.1.1.7+ E484K clades in the United States, with Pennsylvanian isolates in two different clades.
Results showed B.1.1.7 isolates belonged up to seven out of ten different clonal complexes in the lineage. And some of these clonal complexes without the mutation were present since at least November 2020.
Of 235 U.S. isolates, researchers traced the B.1.1.7 + E484K mutation back to three different clades. The genome showed six came from several American states, 18 from Sweden, 2 from Poland, and 1 from Germany. Four isolates from the Pennsylvania area belonged to a large clade with ties to other genomes and a nearby subclade in West Virginia.
The researchers also looked at single nucleotide polymorphisms (SNPs) of 236 isolates, where they found 12 out of 17 had SNPs related to the B.1.1.7 lineage. In addition, one Pennsylvania isolate had all 17 single nucleotide polymorphisms and commonalities with nine other U.S. isolates with a stop mutation in ORF8.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Moustafa AM, Comparative Analysis of Emerging B.1.1.7+E484K SARS-CoV-2 isolates from Pennsylvania. bioRxiv, 2021. doi: https://doi.org/10.1101/2021.04.21.440801, https://www.biorxiv.org/content/10.1101/2021.04.21.440801v1
- Peer reviewed and published scientific report.
Moustafa, Ahmed M, Colleen Bianco, Lidiya Denu, Azad Ahmed, Susan E Coffin, Brandy Neide, John Everett, et al. 2021. “Comparative Analysis of Emerging B.1.1.7+E484K SARS-CoV-2 Isolates.” Open Forum Infectious Diseases 8 (7). https://doi.org/10.1093/ofid/ofab300. https://academic.oup.com/ofid/article/8/7/ofab300/6292249.