
 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Role of Nsp14 in SARS-CoV-2 replication
Nsp14 is a component of the replication-transcription complex with two conserved domains that serve different activities. The N-terminal domain (NTD) is an exoribonuclease (ExoN) that works from 3' to 5', whereas the C-terminal domain is a ribonucleic acid (RNA) cap guanine N7-methyltransferase (MTase) that works from 3' to 5'.
During RNA replication, the N-terminal ExoN domain performs proofreading, thus allowing mismatched nucleotides introduced by the viral RNA polymerase to be removed. The C-terminal MTase domain of Nsp14 is an S-adenosyl methionine (SAM)-dependent methyltransferase that methylates the 5′ guanine of the Gppp-RNA cap at the N7 position, which is required for viral RNA capping.
The 5′ cap is crucial for viral messenger RNA (mRNA) stability and translation, as well as for evading innate antiviral responses in the host. Nsp14 causes translational halting, participates in innate immunity evasion, activates proinflammatory signals, and orchestrates viral recombination.
The Nsp14 protein of SARS-CoV-2 interacts with SIRT5. Sirtuins are key regulators of cellular metabolism and aging that use nicotinamide adenine dinucleotide (NAD) as a co-substrate.
SIRT5 binds stably with non-structural protein (Nsp14) but not with its cofactor Nsp10, and the enzymatic activity of SIRT5 is required for the association. The knocking out or inhibiting of SIRT5 in cell culture tests has lowered viral loads, thereby indicating that SIRT5 is a proviral component required for successful viral replication.
SARS-CoV-2 hijacks the cellular machinery after entering the cell, which allows for its viral proteins to physically interact with hundreds of human proteins. SIRT5 has been identified as a potential binding partner of Nsp14 in protein interactome studies. The nature of this interaction and the role of SIRT5 during SARS-CoV-2 infection were examined in this study.
Study findings
The findings of the current study showed that SIRT5 interacts with Nsp14 in a stable manner and that this connection is independent of Nsp10. Interestingly, the researchers discovered that the catalytic activity of SIRT5 was required for the interaction, as many SIRT5 catalytic mutants were unable to bind to Nsp14 and certain SIRT5 inhibitors prevented the association.
Although SIRT5 is the primary cellular desuccinylase, demalonylase, and deglutarylase, the researchers were unable to find these lysine modifications on the Nsp14 protein, thus implying that SIRT5 does not modify Nsp14 directly. SIRT5 is also a proviral factor, as demonstrated by the fact that when SIRT5 is removed or inhibited in cell-culture trials, SARS-CoV-2 levels drop. The researchers discovered that SIRT5-knockout (KO) cells have greater basal levels of innate immunity markers and produce a stronger antiviral response, which could explain the decrease in virus levels.
The association between SIRT5 and Nsp14 was prevented when the SIRT5 catalytic domain was mutated or when cells were treated with a SIRT5 inhibitor. Furthermore, the strength of the contact appeared to be controlled by cellular NAD levels.
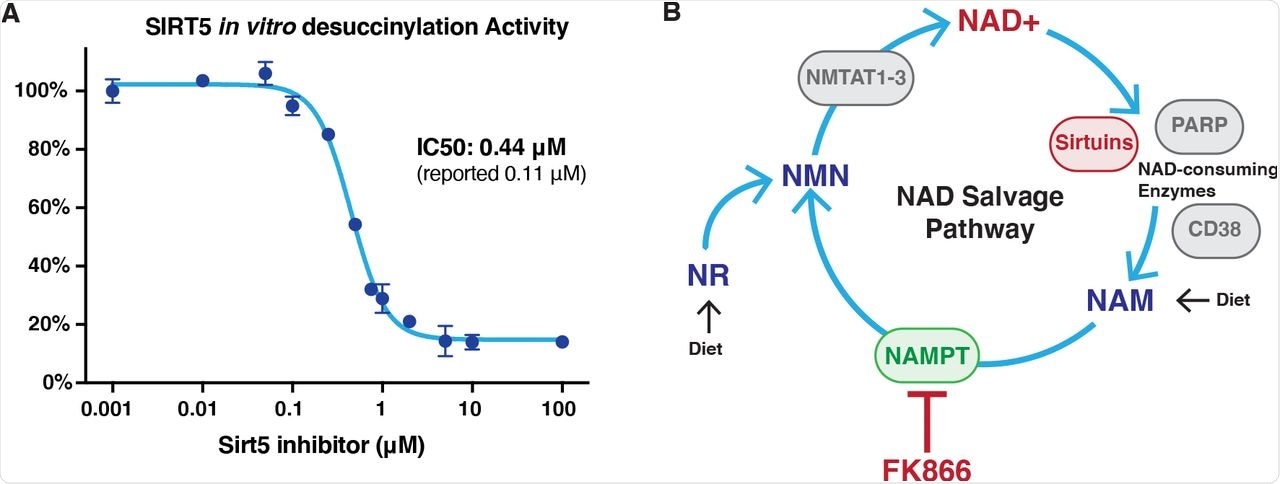
Characterization of inhibitors.
Using mass spectrometry and immunoblotting, the researchers observed no lysine changes on Nsp14. Furthermore, SIRT5 is the only known desuccinylase, demalonylase, and deglutarylase, and these investigations were carried out in Sirt5-KD cells, which would have enriched the presence of these lysine modifications if they existed.
SIRT5-KO mice have no noticeable phenotype and have strong innate immune responses to a variety of bacterial infections. Even in mock-infected cells, the reduction in SARS-CoV-2 levels was linked with enhanced basal levels of many viral restriction factors in SIRT5-KO cells.
This small but considerable increase could explain the slow propagation of SARS-CoV-2 in SIRT5-KO cells. The findings also suggested that SIRT5 desuccinylation of mitochondrial antiviral signaling protein (MAVS) is not the primary mechanism causing viral levels to drop in SIRT5-KO cells.
Conclusions
Further research into the potential link between, the Nsp14/SIRT5 interaction and the proviral role of SHIRT5 needs to be conducted.
There are several hypotheses to consider. First, Nsp14 could promote viral replication by increasing SIRT5 activity, which would decrease innate immune responses and favor viral replication. Second, SIRT5 could be redirected by Nsp14 to other targets, such as other viral proteins.
Third, as noted by the researchers, SIRT5 and Nsp10 formed distinct complexes and SIRT5 boosted Nsp14 MTase activity marginally. The absence of SIRT5 causes a deficiency in cap methylation, more efficient immunological detection of viral RNA, and greater immune response.
Taken together, SIRT5 is a possible pharmacological target that could help fight viral infections in general and SARS-CoV-2 in particular. Inhibiting SIRT5 will certainly never be utilized as the first line of defense; however, it could be used along with medications that directly target viral enzymes to create innovative COVID-19 treatment regimens.
We further showed that SIRT5 is a proviral factor and that SARS-CoV-2 levels decrease when SIRT5 is deleted or inhibited in cell-culture experiments.”

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Walter, M., Chen, I. P., Vallejo-Gracia, A., et al. (2022). SIRT5 is a proviral factor that interacts with SARS-CoV-2 Nsp14 protein. bioRxiv. doi:10.1101/2022.01.04.474979. https://www.biorxiv.org/content/10.1101/2022.01.04.474979v1.
- Peer reviewed and published scientific report.
Walter, Marius, Irene P. Chen, Albert Vallejo-Gracia, Ik-Jung Kim, Olga Bielska, Victor L. Lam, Jennifer M. Hayashi, et al. 2022. “SIRT5 Is a Proviral Factor That Interacts with SARS-CoV-2 Nsp14 Protein.” Edited by Meike Dittmann. PLOS Pathogens 18 (9): e1010811. https://doi.org/10.1371/journal.ppat.1010811. https://journals.plos.org/plospathogens/article?id=10.1371/journal.ppat.1010811.