Throughout the coronavirus disease 2019 (COVID-19) pandemic, scientists have worked at an unprecedented rate to design and develop therapeutics and vaccines to prevent the severe effects of infection with the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2).

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
Remdesivir (RDV) has been one of the few antiviral drugs that received emergency approval for the treatment of SAR-CoV-2 infection. RDV is a ribonucleic acid (RNA) polymerase (nsp12) inhibitor of several viral families, such as Coronaviridae, Filoviridae, Pneumoviridae, and Paramyxoviridae.
Currently, scientists are in the process of characterizing the clinical resistance developed against RDV. To date, there has been a limited amount of documented evidence regarding the in vitro resistance of this antiviral compound against SARS-CoV-2 and other viruses.
In a new study published on the bioRxiv* preprint server, researchers select and study an RDV-resistant virus by serial passaging of a wild type (WT) SARS-CoV-2 Canadian isolate in the presence of increasing concentrations of RDV in cell culture.
Study findings
In the current study, scientists identified V166L substitution in the nsp12 protein after nine passages reaching 6 micromolar (µM) of RDV. The authors revealed that this mutation is not present in the wild-type (WT) SARS-CoV-2 passaged in similar conditions in the absence of RDV drugs.
Development of tiny plaques or absence of cytopathic effects were observed when isolates were further passaged beyond 10 µM of RDV. The scientists performed a plaque reduction assay in Vero E6 cells and determined that virus isolates from passages six, ten, and seventeen demonstrated a reduction of susceptibility by 3.9-, 2.7-, and 2.3-fold to RDV, respectively, as compared to the WT isolate.
During the entire RDV-resistant selection process, the scientists found that the V166L substitution has been the only mutation found outside of the polymerase active site of the nsp12 protein. However, contacts residues were observed in the active site Motifs A and D after nine passages.
Modeling studies revealed that V166L provides only a meek perturbation of Motif D. Therefore, any impact from this substitution on RDV potency is suspected to be indirect. The researchers also reported that V166L remained the only nsp12 substitution after seventeen passages at a final concentration of 10 µM RDV.
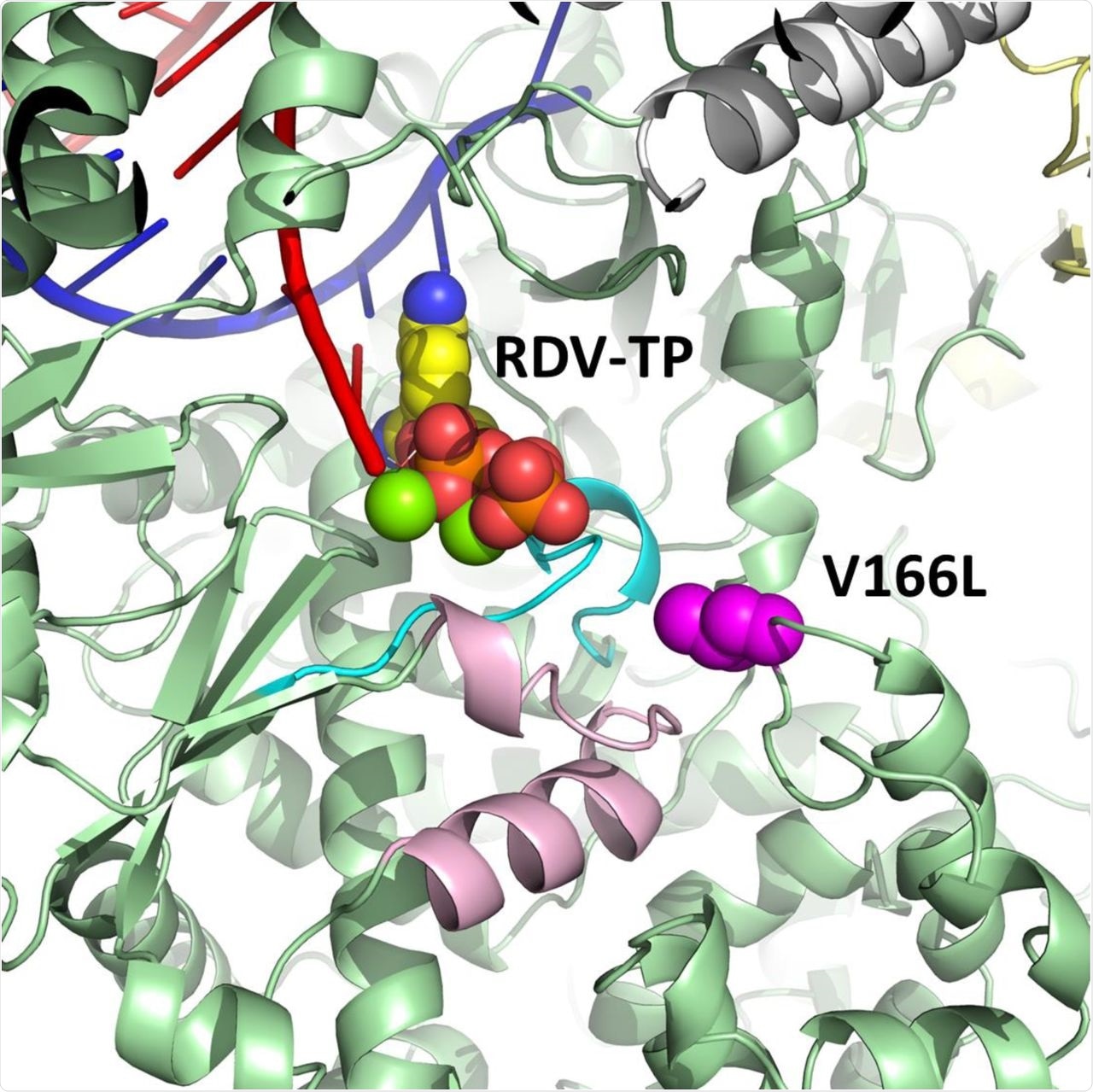
Model of pre-incorporated remdesivir triphosphate (RDV-TP) with a V166L substitution (magenta) in SARS-CoV-2 nsp12 (green).
After the introduction of V166L into a SARS-CoV-2-Nluc recombinant WT WA1 virus, a 1.5-fold increase in the half-maximal effective concentration (EC50) of RDV was observed through a luciferase assay. This finding suggests a high in vitro barrier to RDV resistance.
An extensive screening in the sequencing databases in January 2022 showed the presence of V166L substitution in 0.004% of SARS-CoV-2 sequences present.
In an immunocompromised patient with prolonged SARS-CoV-2 shedding, the V166L substitution in nsp12 was found only after treatment with two 10-day courses of RDV and two units of convalescent plasma. This observation was is in line with previous studies associated with other coronaviruses that reported substitution caused resistance to RVD.
A previous study based on the passage of the murine hepatitis virus (MHV) in the presence of GS-441524 (parent nucleoside analog of RDV), selected two substitutions of F476L and V553L, which caused a five-fold reduction of susceptibility to RDV. Similar results were obtained in an in vitro study using a mouse model, where the corresponding mutation resulted in a six-fold decrease in the efficacy of the RDV drug.
Conclusions
The introduction of the V166L mutation in the recombinant virus caused a 1.5-times increase in the RDV EC50 value. The emergence of only a single V166L substitution, after an increased number of passages and minimal alteration to RVD susceptibility, might be due to a high genetic barrier of resistance for RDV.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Checkmahomed, L., Du Pont, V., Riola, N. C., et al. (2022) In Vitro Selection of Remdesivir-Resistant SARS-CoV-2 Demonstrates High Barrier to Resistance. bioRxiv. doi:10.1101/2022.02.07.479493. https://www.biorxiv.org/content/10.1101/2022.02.07.479493v1.
- Peer reviewed and published scientific report.
Checkmahomed, Liva, Julie Carbonneau, Venice Du Pont, Nicholas C. Riola, Jason K. Perry, Jiani Li, Bastien Paré, et al. 2022. “In Vitro Selection of Remdesivir-Resistant SARS-CoV-2 Demonstrates High Barrier to Resistance.” Antimicrobial Agents and Chemotherapy 66 (7). https://doi.org/10.1128/aac.00198-22. https://journals.asm.org/doi/10.1128/aac.00198-22.