Although several studies have focused on the mechanism of immune evasion by SARS-CoV-2, further research is still needed to understand how the virus modulates the release of type I interferons (IFNs).
 Study: SARS-CoV-2 type I Interferon modulation by nonstructural proteins 1 and 2. Image Credit: NIAID
Study: SARS-CoV-2 type I Interferon modulation by nonstructural proteins 1 and 2. Image Credit: NIAID

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
About the study
In the present study, researchers characterized the response of type I IFN during SARS-CoV-2 infection and the immune evasion mechanisms used by the virus.
The team studied the innate immune response against the SARS-CoV-2 Wuhan strain and the SARS-CoV-2 Beta and Delta variants by infecting K18-human angiotensin-converting enzyme 2 (hACE2) mice with a lethal dose of SARS-CoV-2. Furthermore, the expression of genes involved in the nucleotide-binding and oligomerization domain (NOD)-like receptors (NLRs), toll-Like receptors (TLRs), and retinoic acid-inducible gene (RIG)-like receptors (RLR) signaling pathways were estimated. The team also monitored the expression of genes associated with the type 1 IFN signaling during infection.
Furthermore, the researchers infected A549-hACE2 with the Wuhan strain, followed by visualization via immunofluorescence using anti-SARS-CoV-2 nucleocapsid (N) antibodies. Subsequently, IFN-beta messenger ribonucleic acid (mRNA) was quantified using quantitative reverse transcription-polymerase chain reaction (RT-qPCR). The team also explored the behavior of non-structural protein 1 (nsp1) in the absence of nsp2 by transfecting expression vectors associated with nsp1 and nsp2 into HEK293T cells.
Results
The study results showed that the copy number of the SARS-CoV-2 envelope (E) gene or genomic RNA was almost three times higher in the SARS-CoV-2 Beta-infected mice as opposed to the Wuhan-infected ones. Moreover, the viral RNA loads in the Delta-infected mice were higher than in the Wuhan-infected mice. Similar viral loads were also observed among lung homogenates derived from the Wuhan, Beta, and Delta-infected groups of mice.
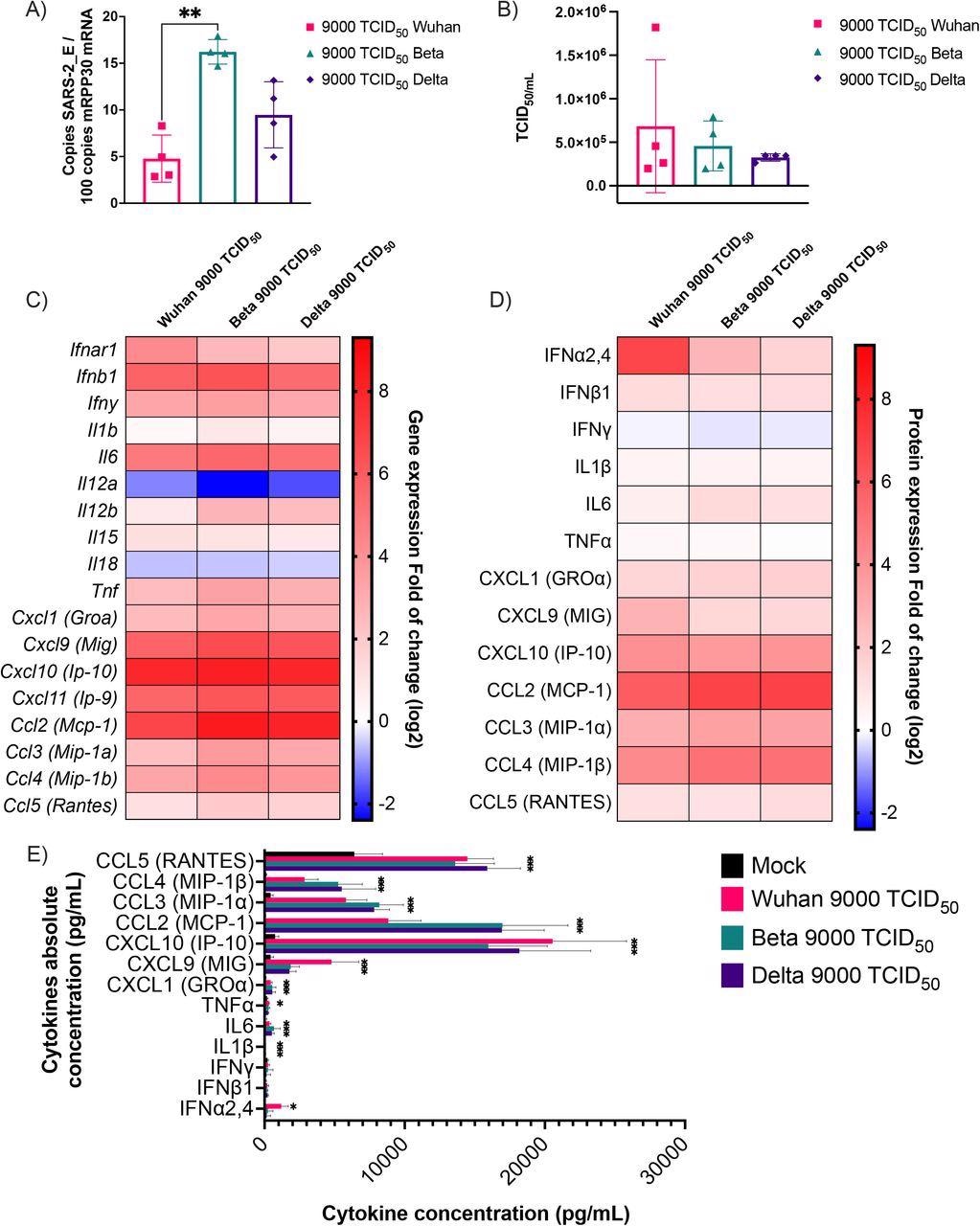
Cytokine mRNA and protein expression profile following infection of K18-ACE2 mice with Wuhan, Beta and Delta strains. Infected or mock mouse lung tissues were collected three days post-infection (n=4/group). A) The number of SARS-CoV-2 E gene copy number was evaluated by ddPCR using lung RNA and expressed as copie number per 100 copies of Rpp30 mRNA. (B) Infectious viral titers were determined in lung homogenates and expressed in TCID50/mL. (C-D) Gene expression was evaluated by RT-qPCR and cytokine concentration in lung homogenates determined using a 13-plex Luminex panel. Cytokine gene expression and concentration levels are presented as heatmaps with results expressed as fold (log2) relative to mock-infected mice. Statistical analyses were done by comparing 2(- ΔCt) values for each gene in the control group and infected groups with a nonparametric T-test and only data with a p value less than 0,05 are show. (E) Absolute cytokine concentrations in lung homogenates. Results are expressed as mean +/-SD (n=4 mice/group). For protein quantification, statistical analyses were done by comparing the normalized concentration for each cytokine in the control group and infected groups with a nonparametric T-test. *P<0,05, **P<0,01, ***P<0,001, ****P<0,0001.
The primary cytokine genes that were upregulated by all the viral variants were chemokine C-X-C ligand 9 (Cxcl9), Cxcl10, Cxcl11, chemokine C-C ligand 2 (Ccl2), interleukin 6 (IL-6), and IFNb1. On the other hand, IL11a and IL18 were downregulated. In addition, the team noted that CC and CXC chemokines were sufficiently induced after viral infection. Moreover, the Wuhan strain displayed high induction of gene expression leading to the release of IFN-alpha, while the Beta and Delta variants exhibited no such induction.
Additionally, the production of chemokines was 1,000 to 2,000 times higher compared to that of pro-inflammatory cytokines and type I IFNs. Overall, this showed that the innate immune response in mice against SARS-CoV-2 was driven by chemokine production.
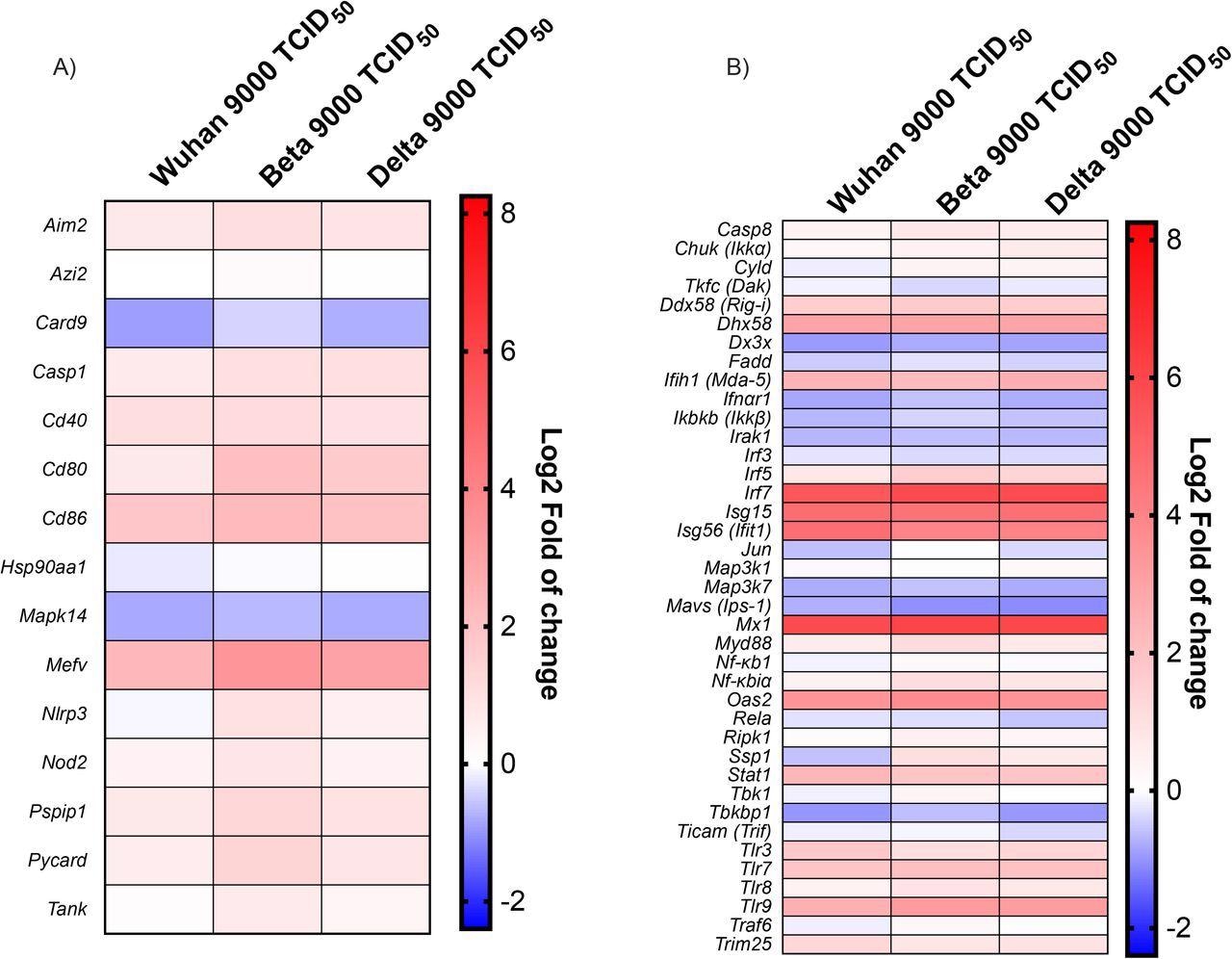
Antiviral response gene expression following infection with Wuhan, Beta and Delta strains. Heat map representation of cytokines and inflammatory-related genes (A) and Type I IFN production and signalization-related genes (B). Results are expressed as fold (log2) relative to mock-infected mice. For gene expression, statistical analyses were done by comparing 2(- ΔCt) values for each gene in the control group and infected groups with a nonparametric T-test, and only data with p values less than 0,05 were shown. For protein expression, statistical analyses were done by comparing the normalized concentration of each cytokine in the mock-infected groups with infected groups with a nonparametric T-test. *P<0,05, **P<0,01, ***P<0,001, ****P<0,0001.
The team noted that the formation of inflammasomes and the release of pro-inflammatory cytokines implied the significant upregulation of the Mediterranean fever gene (Mefv). However, inflammasome components, including apoptosis-associated speck-like protein containing a CARD (Pycard), absent in melanoma 256 2 (Aim2), proline-serine-threonine phosphatase-interacting protein 1 (Pspip1), NLR family pyrin domain containing 3 (Nlrp3), and Caspase 1 (Casp1) were not modulated in the initial phases of the infection. Furthermore, mitogen-activated protein kinase 14 (Mapk14) and Caspase recruitment domain-containing protein 9 (Card9) were downregulated as a response to a SARS-CoV-2 infection.
Several downstream effector genes including inhibitor of nuclear factor kappa-B kinase subunit beta (Ikbkb), transcription factor, interleukin-1 receptor-associated kinase 1 (Irak1), and mitogen-activated protein kinase kinase kinase 7 (Map3k7) were downregulated during SARS-CoV-2 infection.
SARS-CoV-2 infection of A549-hACE2 showed that SARS-CoV-2 efficiently stimulated the transcription of the IFN-beta 1 gene. However, the team found no production of IFN-beta 1 protein in the supernatant of the cells infected with SARS-CoV-2. Moreover, SARS-CoV-2 infected cells effectively produced a limited amount of the IFN-beta 1 protein, which suggested the impact of viral factors on mRNA translation. The team also noted that nsp1 significantly inhibited the Sendai virus (SeV)-induced activation of the IFN-beta 1 promoter. On the other hand, nsp2 expression stimulated IFN-beta 1 as well as interferon-sensitive response element (ISRE) promoters and also amplified its response to IFN-alpha and SeV.
The team also observed that nsp1 and nsp2 did not impact the transcription of IFN-beta 1, interferon-stimulated gene 15 (ISG15), and ISG56. In contrast, nsp1 substantially inhibited the production of IFN-beta 1, while the presence of nsp2 partially reduced this inhibition. Altogether, nsp-1 decreased the IFN response by inhibiting mRNA translation.
Overall, the study findings showed that SARS-CoV-2 stimulated vigorous expression of inflammatory as well as antiviral genes. The researchers believe that nsp2 could be a potential target for future therapeutic approaches against SARS-CoV-2 pathogenesis.

 *Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
*Important notice: bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
Journal reference:
- Preliminary scientific report.
SARS-CoV-2 type I Interferon modulation by non structural proteins 1 and 2, Louis Flamand, Émile Lacasse, Isabelle Dubuc, Isabelle Allaeys, Leslie Gudimard, Annie Gravel, Eric Boilard, bioRxiv 2022.06.09.495586, DOI: https://doi.org/10.1101/2022.06.09.495586, https://www.biorxiv.org/content/10.1101/2022.06.09.495586v1