A new study shows that gut microbiome signatures, analyzed through advanced machine learning, can help identify individuals with more severe insulin resistance, offering fresh insight into the biological links between dysbiosis and metabolic dysfunction.
Study: Exploring the gut microbiome in type 2 diabetes across different insulin resistance levels: a machine learning approach. Image Credit: Kateryna Kon / Shutterstock
In a recent study published in the journal Frontiers in Nutrition, researchers investigated associations between insulin resistance (IR), a primary driver of type 2 diabetes mellitus (T2DM), and gut microbiome (GM) composition. Using stool-derived 16S ribosomal RNA (rRNA) gene sequencing and blood-based metabolic markers from 116 participants, the team trained machine learning (ML) models to determine whether gut microbial signatures could distinguish individuals with elevated insulin resistance from healthy controls.
The study found that extreme gradient boosting (XGBoost) models could differentiate individuals with higher insulin resistance from controls with moderate accuracy. These findings suggest that targeting specific gut bacterial groups may represent a future adjunctive intervention strategy to improve metabolic regulation in patients with T2DM, pending further validation.
Insulin Resistance and the Gut Microbiome in Type 2 Diabetes
T2DM is a chronic metabolic disorder characterized by persistent hyperglycemia, which significantly increases the risk of cardiovascular disease and other serious metabolic complications if left untreated. IR is widely recognized as the principal pathophysiological driver of T2DM. It occurs when peripheral tissues, including muscle and liver, fail to respond effectively to insulin, disrupting carbohydrate and lipid metabolism.
Although pharmacological therapies such as metformin target these metabolic pathways, growing evidence suggests that the GM may function as a complementary therapeutic target rather than a superior alternative. Consequently, research increasingly aims to clarify microbiome–insulin resistance interactions and identify key bacterial communities linked to metabolic dysfunction.
Microbiome data are inherently high-dimensional and complex, making it difficult to identify specific microbes associated with varying IR severity using conventional computational approaches. Advances in ML enable the integration and analysis of large, multimodal datasets, revealing patterns that are not easily detected by traditional statistical methods.
Study Design: 16S rRNA Sequencing and ML-Based Classification
The study enrolled 116 participants from Chengdu, China, including 78 individuals with clinically diagnosed T2DM and 38 healthy controls. Blood samples were collected for metabolic profiling, while stool samples underwent 16S rRNA gene sequencing to characterize gut microbiome composition.
Metabolic status was assessed using standard clinical biomarkers, including fasting blood glucose (FBG), triglycerides (TG), and high-density lipoprotein cholesterol (HDL-C). Because direct measurement of insulin resistance is clinically challenging, researchers calculated four validated composite indices to estimate IR severity.
- Atherogenic index of plasma (AIP).
- Metabolic score for insulin resistance (METS-IR).
- Triglyceride-glucose index (TyG).
- TyG-body mass index (TyG-BMI).
XGBoost models were trained to determine whether gut microbiome signatures could classify individuals with high insulin resistance relative to controls. Feature-importance analyses identified bacterial taxa most strongly associated with metabolic dysfunction.
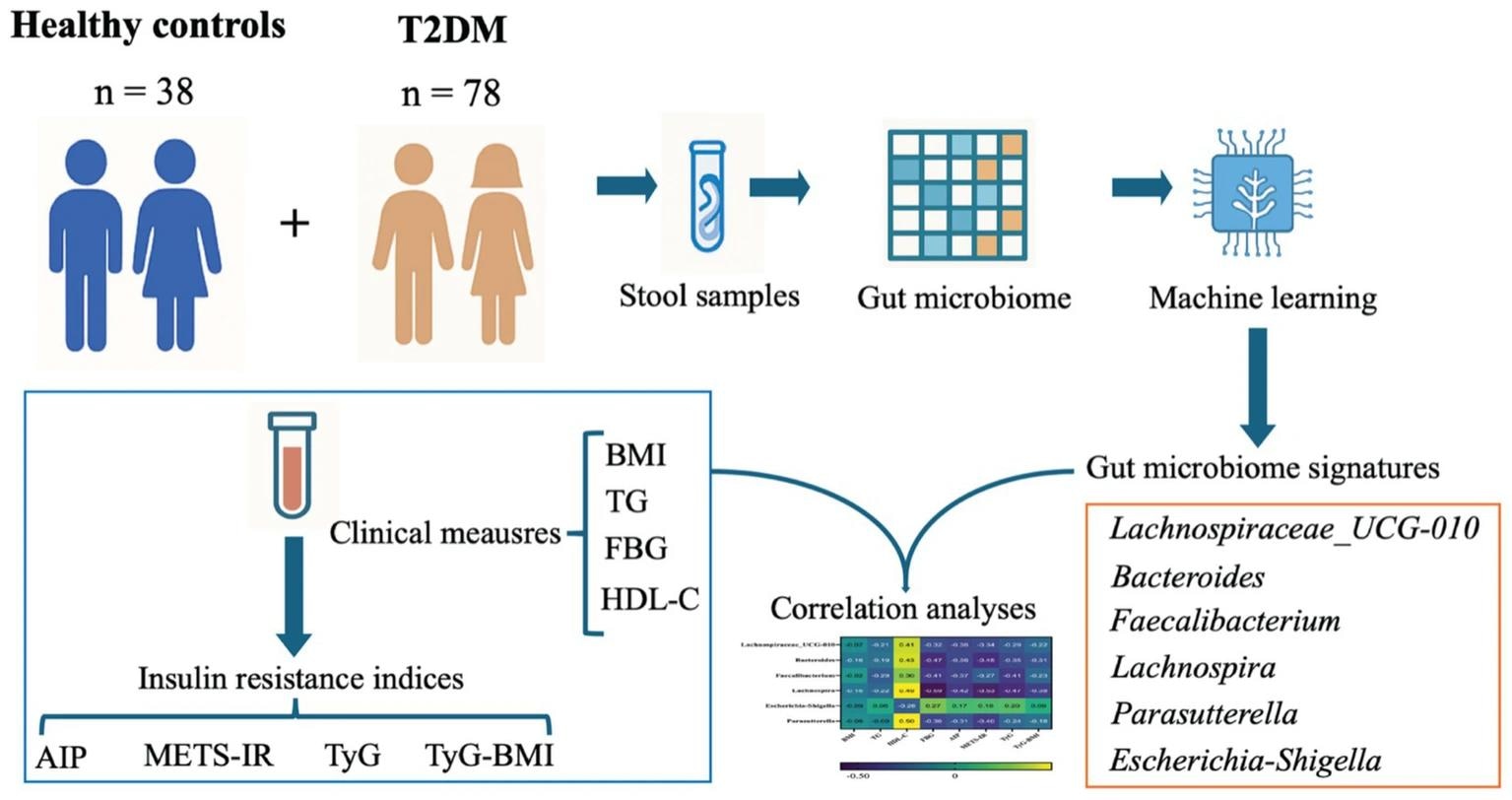
Graphical abstract. Machine-learning-based identification of gut microbiome taxa associated with insulin resistance (IR) in type 2 diabetes. A cohort of healthy controls and T2DM participants provided stool samples for gut microbiome profiling. In parallel, clinical measurements including BMI, TG, FBG, and HDL-C were collected and used to derive IR indices including AIP, METS-IR, TyG, and TyG-BMI, to capture complementary dimensions of glucose-lipid dysregulation. Microbiome features were then selected using a machine learning framework, yielding a set of IR-related microbial features. Finally, correlation analyses integrated the selected taxa with clinical and IR indices to nominate microbial features potentially relevant to IR modulation.
Metabolic Differences Between T2DM and Controls
Although body mass index (BMI) did not significantly differ between T2DM patients and controls (25.48 vs. 24.84 kg/m2, p > 0.05), substantial metabolic differences were observed. Individuals with T2DM exhibited elevated TG (2.60 vs. 1.49 mmol/L, p < 0.0001) and FBG (8.32 vs. 5.10 mmol/L, p < 0.0001), alongside reduced HDL-C levels (0.91 vs. 1.27 mmol/L, p < 0.0001). All four calculated IR indices were significantly higher in the diabetic cohort (p < 0.0001), confirming a pronounced metabolic divide between groups.
Machine Learning Performance and AUC Results
The XGBoost models demonstrated moderate ability to identify severe insulin resistance using gut microbiome sequencing data alone. The METS-IR–based classifier achieved the strongest performance, with an area under the receiver operating characteristic curve (AUC) of 0.84. While promising, this level of discrimination does not meet diagnostic thresholds but does support the feasibility of microbiome-informed metabolic risk stratification.
Microbial Shifts Associated With Insulin Resistance
Feature-importance analysis revealed distinct microbial alterations linked to metabolic dysfunction. Beneficial short-chain fatty acid–producing bacteria were significantly reduced in individuals with T2DM. For example, the relative abundance of Bacteroides was 9.39% in diabetic patients compared to 25.33% in controls.
Potentially pathogenic bacteria such as Escherichia-Shigella were significantly elevated in the T2DM group (8.48% vs. 1.97%). These compositional shifts correlated with glycemic and lipid abnormalities, although the cross-sectional design precludes conclusions regarding causality.
Clinical Implications, Limitations, and Future Directions
This study highlights the potential utility of ML models in identifying gut microbiome features associated with insulin resistance and metabolic dysfunction. Specific taxa, including Faecalibacterium and Escherichia-Shigella, may serve as microbial signatures associated with disease severity.
Limitations include the cross-sectional design, modest sample size, potential confounding factors such as diet, medication use, and lifestyle variables, and limited taxonomic resolution inherent to 16S rRNA sequencing. Despite these constraints, the findings demonstrate that distinct gut microbiome profiles are associated with insulin resistance and disruption of glucose and lipid metabolism characteristic of T2DM.
Future longitudinal and interventional studies are needed to determine causality and assess whether personalized probiotics, microbiome-targeted dietary strategies, or other microbiome-modulating interventions can serve as adjunctive therapies for T2DM rather than stand-alone treatments, pending rigorous clinical validation.