A transposable element (TE) is a DNA sequence that is able to change relative position in the genome, and they in fact make up around 50% of the human genome. Most TEs appear to have no effect on the phenotype, though, depending on where they are inserted into the genome, they may cause mutations to arise. They are thought to be largely involved in homeostasis, and have been demonstrated to be involved in the generation of interferon in response to viral threat.
A study by researchers in Israel aims to establish whether TE expression is altered in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infected cells, and explores the implications for host immune response.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The research team's findings have been released on the bioRxiv* preprint server.
How are transposable elements involved in immune response?
TEs are involved in the transcriptional network that results in interferon response by a diverse range of overlapping and amplifying mechanisms. Additionally, TEs have been noted to be upregulated in T-lymphocyte enhancers, and throughout the whole genome in humans in response to some viruses. Avian flu, for example, has been found to elicit TE activity and subsequently enhance the host immune response.
Interestingly, early experiments with SARS-CoV-2 indicated that TE expression was heightened in A549 adenocarcinomic human alveolar basal epithelial cells, but not in NHBE primary lung epithelial cells, even with similar viral loads. To test this phenomenon further, the group infected stem cell-derived alveolar type 2 (iAT2) cells with SARS-CoV-2, also noting only a mild TE response, though greater than that observed in NHBE. In all cases, the population of TEs was seen to rise before interferon upregulation was detected, indicating a causal relationship.
Several genes were found to correlate with TE expression, with mitochondrial-related processes doing so inversely, while those involved in pre-mRNA processing and transcription were enhanced. Importantly, many genes that produce proteins that interact with SARS-CoV-2 were noted to inversely correlate with TE expression, suggesting that the virus is mediating the host TE response.
Epigenetic alterations to the genome before and after SARS-CoV-2 or avian flu infection were analyzed in A549 cells. TEs generated by cells exposed to SARS-CoV-2 exhibited a distinct epigenetic profile that differed from those generated by avian flu-infected cells, specifically, being devoid of H3K27me3, an epigenetic modification that is associated with the downregulation of nearby genes. This epigenetic aberration, among others observed by the group, renders the cell prone to TE disorder.
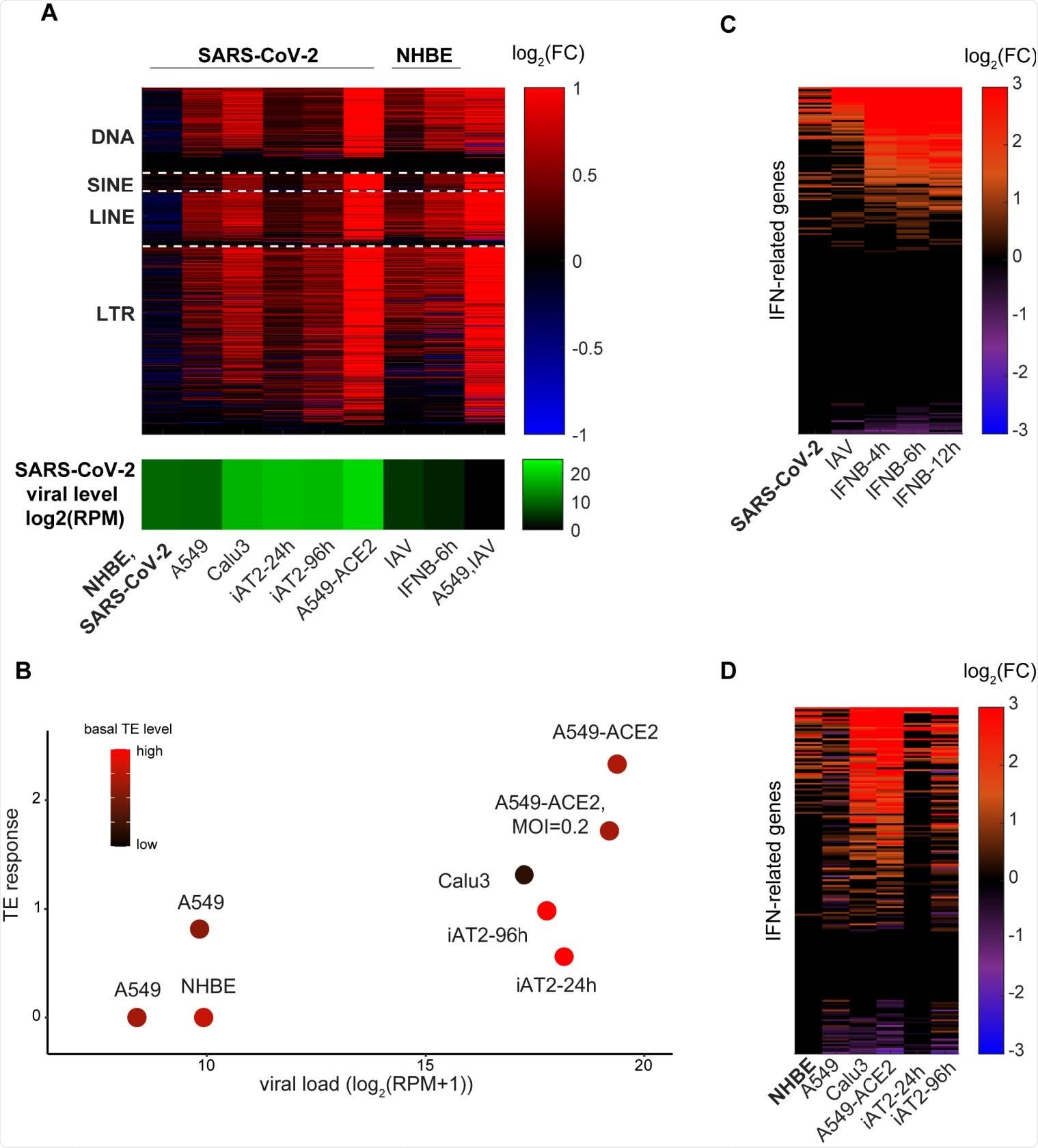
TEs expression changes and IFN response to SARS-CoV-2 and IAV infections. A. Log2-fold change in expression level of TE subfamilies (DNA, SINE, LINE and LTR) in different cell lines and in the different conditions indicated. SARS-CoV-2 viral levels (green) are depicted in the bottom panel. B. TE induction levels is correlated with TE basal levels pre-infection and SARS-CoV-2 viral levels. Linear regression coefficients are 0.158 and -0.57 for viral load and basal TE level, respectively (R2=0.81). C-D. The IFN transcriptional response among NHBE cells (C) and among SARS-CoV-2-infected cells (D).
What are the implications for COVID-19?
A549 cells naturally exhibit lower baseline TE expression rates than iAT2 cells, which are lower still than observed in NHBE cells. Additionally, epigenetic dysregulation of TEs is usually observed in A549 cancer cells, impairing the TE response.
NHBE cells are sourced from a 79-year-old female donor, and TE expression is heightened with age, which is also the major risk factor for COVID-19. The group hypothesizes that TE expression could be a major influencing factor in the association between age and COVID-19 outcome. Those with heightened baseline TE levels experience a proportionately lesser spike in TE expression upon viral infection, resulting in a weaker interferon response. The cells are desensitized towards changes in TE expression.
The group proposes that the impaired TE response observed in SARS-CoV-2 infection results from active evasion mechanisms of the virus, exacerbated by age-resultant epigenetic dysregulation. A weakened initial interferon response could lead to an exaggerated inflammatory response, resulting in a life-threatening cytokine storm.
TE activation could therefore be utilized as a therapeutic tool against COVID-19, generating improved early interferon responses and thus lessening the severity or occurrence of serious inflammatory responses in the host.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources