As of February 22, 2022, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has infected over 425 million worldwide and caused over 5.89 million fatalities worldwide.
The rapid spread of SARS-CoV-2 has enabled the virus to evolve quickly, thereby resulting in the emergence of several variants including the B.1.1.7 (Alpha), B.1.351 (Beta), P.1 (Gamma), B.1.617.2 (Delta), and B.1.1.529 (Omicron) variants. Each of these variants has been designated by the World Health Organization (WHO) as variants of concern (VOCs) as a result of their increased transmissibility, ability to evade immunity, and/or cause severe illness.
Study: An ACE2-blocking antibody confers broad neutralization and protection against Omicron and other SARS-CoV-2 variants. Image Credit: Design_Cells / Shutterstock.com

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Background
SARS-CoV-2 enters host cells through the use of its trimeric spike (S) glycoprotein, which consists of the S1 and S2 subunits. The S1 subunit binds to the angiotensin-converting enzyme 2 (ACE2) receptor present on the surface of host cells, whereas the S2 subunit fuses the membrane.
Neutralizing antibodies primarily target the N-terminal domain (NTD) and the receptor-binding domain (RBD) of the S1 subunit. These domains are common locations for mutations found in SARS-CoV-2 variants that allow serum neutralizing antibodies, as well as NTD- and RBD-directed monoclonal antibodies, to escape from infected or vaccinated patients. More specifically, the four primary and physically characterized neutralizing epitope classes in the RBD are where escape mutations dominantly occur.
In a recent study published on the bioRxiv* preprint server, researchers identify 87G7, a SARS-CoV-2 neutralizing human monoclonal antibody with remarkable broad-spectrum neutralization and protection efficacy. 87G7 inhibits SARS-CoV-2 infection by inhibiting ACE2 binding and has strong neutralizing activity against Alpha, Beta, Gamma, Delta, and Omicron.
Moreover, 87G7 can bind the widely variable ACE2 receptor binding site by targeting a region of hydrophobic residues in the convex edge of the receptor-binding ridge that are highly conserved in SARS-CoV-2 variants, such as the five VOCs, according to structural elucidation.
About the study
The authors examined the antibody repertoire of Harbour H2L2 mice inoculated with the SARS-CoV-2 spike protein to find human monoclonal antibodies with broad neutralizing potential against SARS-CoV-2 VOCs.
Hybridoma supernatants with S ectodomain enzyme-linked immunosorbent assay (ELISA)-reactivity were tested for their neutralizing potential against SARS-CoV-2 S pseudovirus with the S E484K mutation. The E484K mutation differs in multiple SARS-CoV-2 variants of interest (VOIs) and VOCs and plays a crucial role in neutralization resistance.
The 87G7 hybridoma supernatant had the most effective neutralizing activity out of the 300 hybridoma supernatants examined. The authors cloned the human variable heavy and light chain regions into a human immunoglobulin G1 (IgG1)/kappa chain backbone, which allowed for the chimeric 87G7 H2L2 antibody to be converted into a fully human immunoglobulin.
The recombinantly expressed 87G7 human monoclonal antibody (mAb) was tested using the vesicular stomatitis virus (VSV) pseudovirus neutralization assay to determine whether it could neutralize the prototypic Wuhan-Hu-1 SARS-CoV-2 and VOCs.
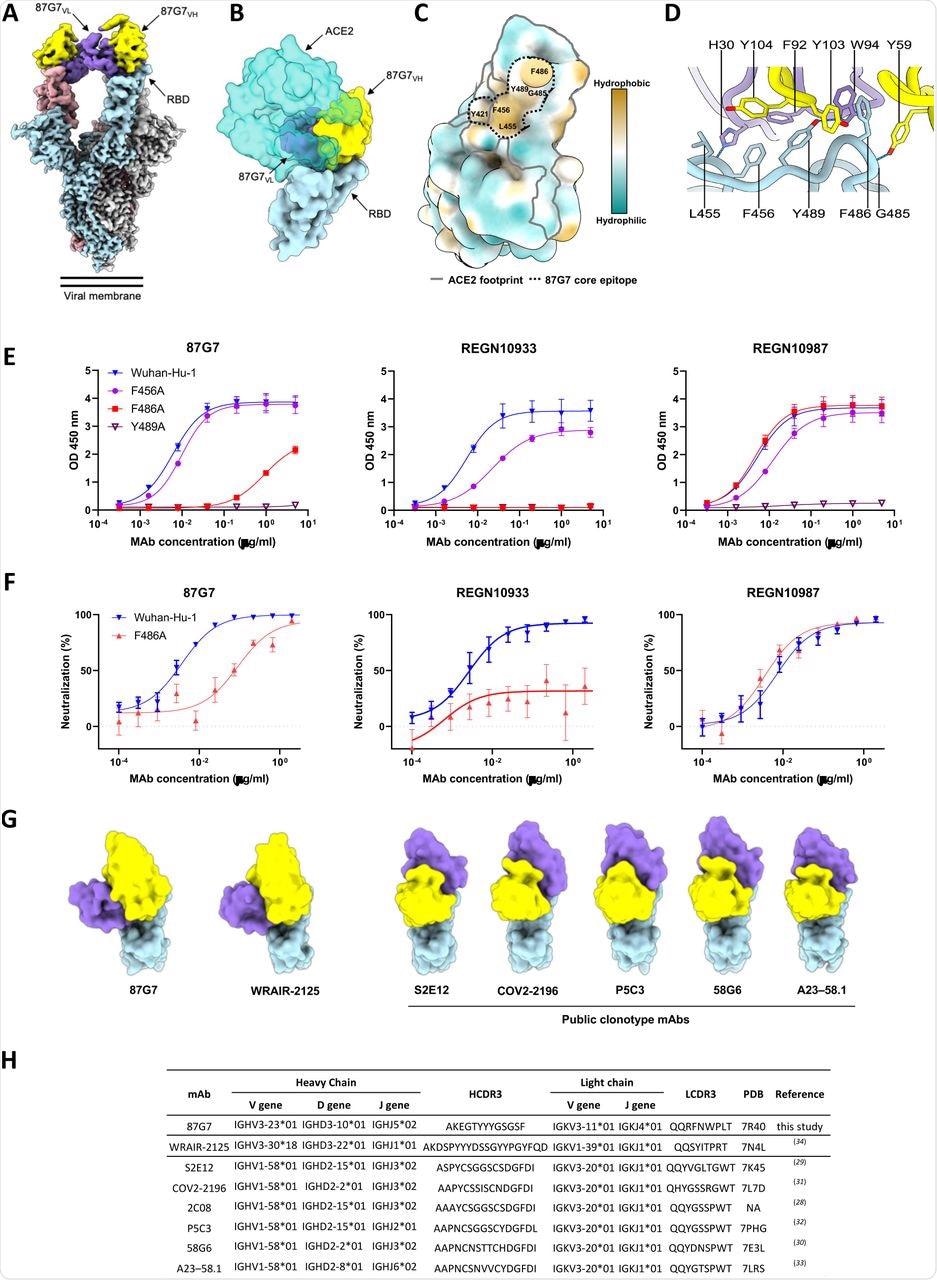
Structural basis for binding and neutralization by 87G7 (A) Composite cryo-EM density map for the SARS-CoV-2 spike ectodomain in complex with the 87G7 antibody Fab fragment. The spike protomers are colored blue, gray, and pink, and the 87G7 light- and heavy-chain variable domains are colored purple and yellow, respectively. (B) Surface representation of the 87G7-bound RBD overlaid with the RBD-bound ACE2 (PDB ID: 6M0J). (C) Surface representation of the RBD colored according to the Kyte-Doolittle scale, where the most hydrophobic residues are colored tan and the most hydrophilic residues are colored blue. The residues which make up the 87G7 core epitope and the ACE2 footprint are outlined. (D) Close-up view showing selected interactions formed between 87G7 and the SARS-CoV-2 RBD (E) ELISA binding of 87G7 to plate-immobilized WT, F456A, F486A and Y489A S1 domains. (F) 87G7 neutralizing activity against pseudoviruses with Wuhan-Hu-1 S and SF486A. REGN10933 and REGN10987 were taken along as a reference in panel E and F. (G) Side-by-side comparison of the SARS-CoV-2 RBD bound to 87G7, WRAIR-2125 (PDB ID: 7N4L), 58G6 (PDB ID: 7E3L), P5C3 (PDB ID: 7PHG), COV2-2196 (PDB ID: 7L7D), S2E12 (PDB ID: 7K45) and A23-58.1 (PDB ID: 7LRS). (H) Germline origins of 87G7 and other F486-directed SARS-CoV-2 mAbs with broad neutralization capacity. NA: not applicable.
Study findings
For comparison, the authors employed two therapeutic mAbs, REGN10933 and REGN10987. With a half-maximal inhibitory concentration (IC50) of 5.4 ng/ml, 87G7 showed significant neutralizing effectiveness against Wuhan-Hu-1 S induced cell entrance.
In addition, with IC50 values ranging from 1.4 to 5.1 ng/ml, entry of VSV pseudotypes containing S proteins from VOCs such as Alpha, Beta, Delta, and Omicron was prevented. The neutralization efficiency of REGN10933 against the SARS-CoV-2 Beta and Omicron variants was reduced by 20-fold and 350-fold, respectively, correlating to a 20-fold and 350-fold drop in IC50, whereas the neutralization capacity of REGN10987 against Omicron was lost.
REGN10933 had lost its neutralization capacity against Omicron BA.1 and had a 7.8- and 15.9-fold reduction in inhibition effect against the SARS-CoV-2 Beta and Gamma variants, respectively, as compared to an earlier pandemic strain with D614G spike mutation (D614G). D614G. The SARS-CoV-2 Alpha, Beta, Gamma, Delta, and Omicron VOCs were powerfully neutralized by 87G7, with IC50 values ranging from 3.1 to 12.5 ng/ml. Furthermore, 87G7 neutralized Lambda and Mu variants with the same potency as D614G.
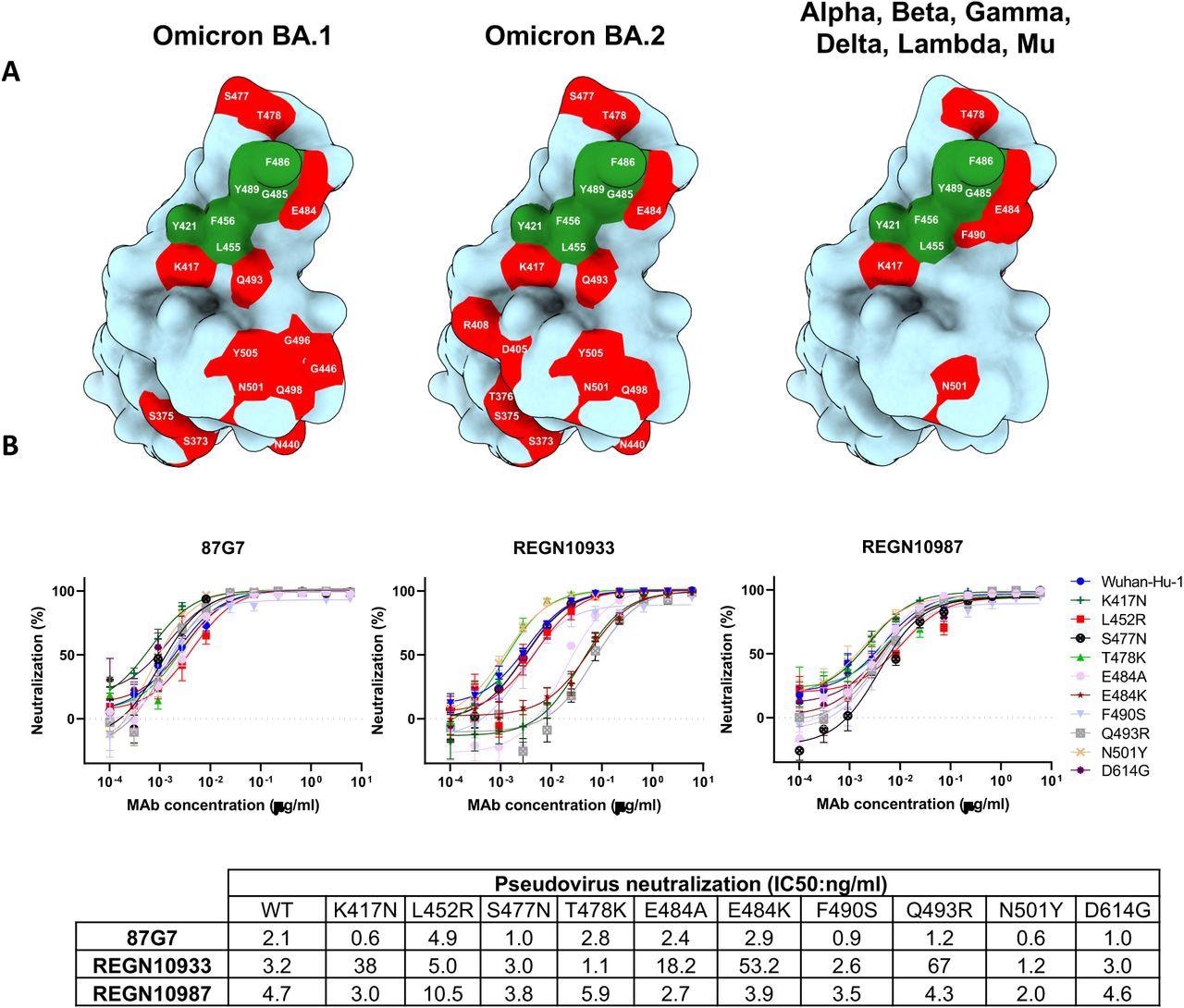
87G7 recognizes a conserved epitope in SARS-CoV-2 RBD (A) Surface representation of the SARS-CoV-2 S RBD with mutations colored red that is found in Omicron BA.1 (left panel) and Omicron BA.2 (middle panel). The right panel displays the set of mutations surrounding the 87G7 core epitope that is present in Alpha, Beta, Gamma, Delta, Lambda or Mu (see also Fig. 1a). The 87G7 core epitope residues are colored green. (B) 87G7 neutralizing activity against pseudoviruses with S variants carrying single residue substitutions found in the SARS-CoV-2 variants of concern. The REGN10933 and REGN10987 therapeutic mAbs were used for benchmarking. Data are shown as mean (± SEM) of two independent experiments with technical triplicates, and corresponding IC50 titers are presented in the lower panel.
The K18-hACE2 transgenic mouse model was used to test the in vivo protective capability of 87G7 against SARS-CoV-2 infection. Mice were given 87G7 or an IgG1 isotype control intraperitoneally and then challenged intranasally 16 hours later with 105 PFU of SARS-CoV-2 utilizing the D614G strain and Alpha, Beta, Gamma, or Delta VOCs.
After two days (Alpha and Gamma), three days (D614G and Beta), or four days (Delta) post-infection, mice in the control group began to lose weight. Comparatively, mice treated with 87G7 appeared to display protection against weight loss following the challenge.
These findings corresponded to a decrease in lung antigen concentrations in these mice on day five after the challenge when compared to isotype-control treated mice. Furthermore, the amount of live virus found in lung homogenates dropped by at least one to three orders of magnitude in 87G7 mice as compared to those treated with the control antibody.
Implications
Clinical success has been achieved in the management of outpatients with mild or moderate coronavirus disease 2019 (COVID-19) using anti-SARS-CoV-2 monoclonal antibodies. However, successful SARS-CoV-2-specific mAb treatment for hospitalized patients has proved elusive. Importantly, the production of these monoclonal antibodies may be useful for both therapeutic and preventive therapy of seronegative individuals, including those who do not produce endogenous antibodies in response to vaccination or infection.
The establishment of monoclonal antibodies for the prevention and treatment of COVID-19 has been complicated by the antigenic evolution of SARS-CoV-2. While neutralization possibilities have been the primary criterion for selecting anti-SARS-CoV-2 antibody candidates for clinical use, the antibody's ability to cross-neutralize by targeting highly conserved sites on the spike protein has become far more important to reduce the threat of antibody escape by future variants.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Du, W., Hurdiss, D. L., Drabek, D., et al. (2022). An ACE2-blocking antibody confers broad neutralization and protection against Omicron and other SARS-CoV-2 variants. bioRxiv. doi:10.1101/2022.02.17.480751. https://www.biorxiv.org/content/10.1101/2022.02.17.480751v1.
- Peer reviewed and published scientific report.
Du, Wenjuan, Daniel L. Hurdiss, Dubravka Drabek, Anna Z. Mykytyn, Franziska K. Kaiser, Mariana González-Hernández, Diego Muñoz-Santos, et al. 2022. “An ACE2-Blocking Antibody Confers Broad Neutralization and Protection against Omicron and Other SARS-CoV-2 Variants of Concern.” Science Immunology, April. https://doi.org/10.1126/sciimmunol.abp9312. https://www.science.org/doi/10.1126/sciimmunol.abp9312.