Sites of O-glycosylation on the spike protein were near N-glycosites, suggesting O-glycans complement N-glycans' effect in immune shielding.
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the cause of the COVID-19 pandemic, has some characteristics that were not seen before in human coronaviruses, such as mutation on the receptor-binding domain (RBD) for high affinity to the human angiotensin-converting enzyme 2 (ACE2) and presence of a furin cleavage site between the two subunits of the spike protein.
Several vaccines have now been approved for use, and many more are under clinical trials. Some vaccines are subunit formulations that use recombinant viral proteins for immunization.
As enveloped viruses move through the secretory pathway, they can acquire host glycosylation. Glycosylation can modify the protein molecules' shape, shielding the amino acids and thus preventing recognition by the host immune system.
Two types of human glycosylation are important for enveloped viruses like the SARS-CoV-2: the N-linked glycosylation and the mucin-type O-linked glycosylation, which are initiated by polypeptide GalNAc transferases (GalNAc-Ts) and modify some amino acids.
Several proteins and expression systems have been used to figure out the structure and post-translational modifications of the SARS-CoV-2 spike protein, and some have also been studied as vaccine candidates. However, it is not easy to predict glycosylation events and the precise locations of O-glycans.
The mapping of O-glycosites requires instruments that can support electron transfer dissociation (ETD) fragmentation. In a new study published on the bioRxiv* preprint server, researchers report their results on whether O-glycosylation patterns affect immunogens for vaccine development.
O-glycosylation patterns on insect and human cell lines
Using a previously developed method by the team for glycosite mapping, they compared O-glycosylation patterns on spike proteins expressed in Drosophila and HEK cells. Using electron transfer collision-induced dissociation (ETciD) and higher energy CID (HCD) MS2 fragmentation, the team identified positions where amino acids were modified. They found ten different glycosites on the spike protein expressed in HEK. Of these, seven were analogous to those on the insect ectodomain.
The O-glycosylation patterns were similar on multimeric ectodomains in the different cell lines. However, the RBD O-glycosylation patterns on the insect monomer and dimer were all different. The authors found that most O-glycosites were present near or just within the N-X-S/T sequons on N-glycosylation. In total, the team identified 25 O-glycosites, of which 16 were located within three amino acids from N-glycosites.
Using software, the team next mapped the O-glycosites they identified onto the completely glycosylated full-length spike protein structure. Most of the O-glycosites were mapped to amino acids that could easily access solvents. For both the cell lines, the O-glycans were found evenly over the exposed surface area, blending with the N-glycans.
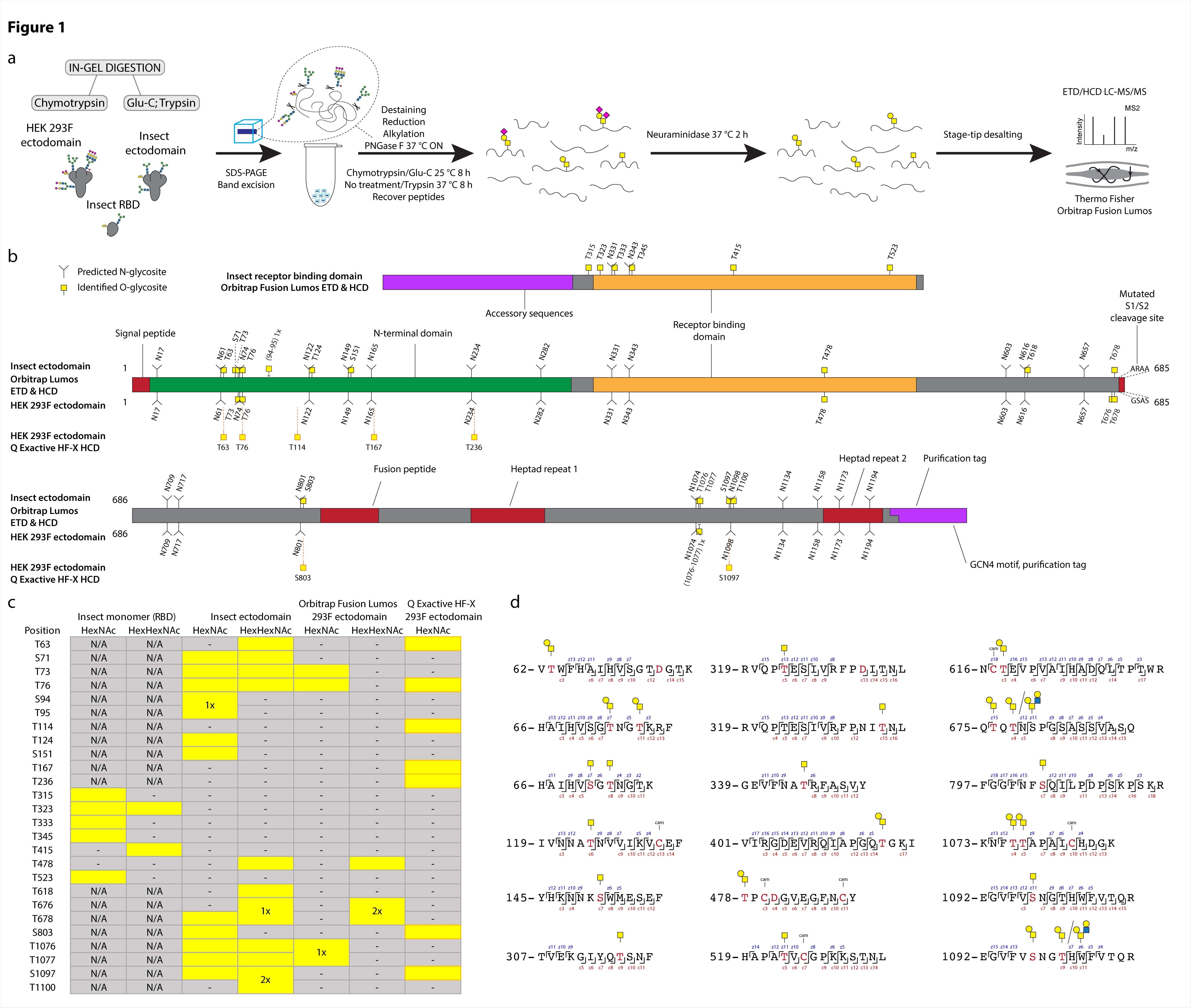
O-glycosylation of SARS-CoV-2 spike protein expressed in insect or human cells. (a) Experimental strategy. (b) A graphical layout of spike protein sequence annotated with identified Oglycosylation sites using Orbitrap Lumos mass spectrometer with ETD and HCD MS2 fragmentation. Independently identified O-glycosites on HEK 293F-derived ectodomain using Q Exactive HF-X mass spectrometer with HCD MS2 fragmentation are shown with orange outlines. (c) A table summarizing O-glycosites and respective structures found in different S formulations (yellow rectangles). Ambiguous sites are shown as merged rectangles across several positions. “N/A” - not applicable; “-” - not detected. Orbitrap Fusion Lumos derived sites are marked with grey outlines. Additional sites identified on HEK 293F ectodomain with Q Exactive HF-X are marked with orange outlines. (d) Examples of O-glycopeptides identified with Orbitrap Fusion Lumos using ETD fragmentation. MS2 c and z product ion fragments are annotated based on the respective ETD spectra
O-glycans likely complement N-glycans in immune shielding

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
The similar glycosylation patterns in human and insect cell lines suggest isoforms of GalNAc-Ts can glycosylate the peptides. The authors found that more than 60% of the O-glycosite sites were next to N-glycosite locations. Further analysis revealed the O-glycosites were located on peptides with asparagine that were not modified by N-glycans. In seven out of nine regions investigated, more than 85% of the peptides not N-glycosylated were O-glycosylated, suggesting regions that were not N-glycosylated was O-glycosylated. It is likely the O-glycans shield peptide fragments that are not being occupied by N-glycans.
The analysis further revealed that O-glycosites have low occupancy, but this low occupancy next to N-glycosites is consistent with published reports on the spike protein N-glycosylation. The low numbers of O-glycosites are unlikely to affect protein function or immunogenicity and are not likely to concern immunogen formulations of highly N-glycosylated protein.
Molecular modeling suggested the N-glycan and O-glycans were often oriented away from each other. It will be useful to understand if the process of formation of mutations on specific sites linked to N-glycans would increase O-glycans in these regions. In addition, investigating the abundance of O-glycans in viruses with less densely populated N-glycosylation will also explain how O-glycosylation affects immune shielding.
Thus, although the O-glycans are a minor part of the spike protein glycan shield, they play an important part in occupying the regions that N-glycosylation does not cover, likely complement the effect of N-glycans in immune shielding.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Bagdonaite, I. et al. (2021). Site-specific O-glycosylation analysis of SARS-CoV-2 spike protein produced in insect and human cells. bioRxiv. https://doi.org/10.1101/2021.02.03.429627, https://www.biorxiv.org/content/10.1101/2021.02.03.429627v2
- Peer reviewed and published scientific report.
Bagdonaite, Ieva, Andrew J. Thompson, Xiaoning Wang, Max Søgaard, Cyrielle Fougeroux, Martin Frank, Jolene K. Diedrich, et al. 2021. “Site-Specific O-Glycosylation Analysis of SARS-CoV-2 Spike Protein Produced in Insect and Human Cells.” Viruses 13 (4): 551. https://doi.org/10.3390/v13040551. https://www.mdpi.com/1999-4915/13/4/551.