Phylogenetics is an analytical tool that quickly analyzes genomic data to provide invaluable insights into the evolution and spread of a pathogen, thereby allowing public health officials and governments to respond to it in a timely fashion.
During the coronavirus disease 2019 (COVID-19) pandemic, phylogenetics, like many other pre-pandemic tools, became redundant owing to the massive scale of genome sequencing data of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) deposited across online databases since 2020.

Study: Pandemic-scale phylogenetics. Image Credit: majcot / Shutterstock.com

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
About the study
In a recent preprint study published on the bioRxiv* server, researchers developed a phylogenetic package that incorporated several pandemic-specific optimization and parallelization techniques. The package comprises four programs UShER, matOptimize, RIPPLES, and matUtils.
To build a comprehensive SARS-CoV-2 phylogeny, SARS-CoV-2 genome sequence data was gathered from major online databases such as the global initiative on sharing all influenza data (GISAID) and GenBank. The GenBank MN908947.3 sequence was used as the reference for rooting the tree and for calling variants in individual samples. In experiments, the sampling date metadata was used to derive two subtrees including a 100K-sample tree and a 1M-sample tree.
All experiments conducted throughout the study were performed on the Google Cloud Platform (GCP) for easy reproducibility. Since this phylogenetic package was memory-efficient, CPU-optimized E2 instances could have been used.
In lieu, memory-optimized instances were used in the package for some competing tools, whereas iso-cost comparison was done to ensure that the hourly cost remained about the same for both instances. Strong and weak scaling analyses were performed for UShER, matOptimize, and RIPPLES using the 1M-sample tree and e2-high CPU-32 instances, varying the number of instances from 2 to 32.
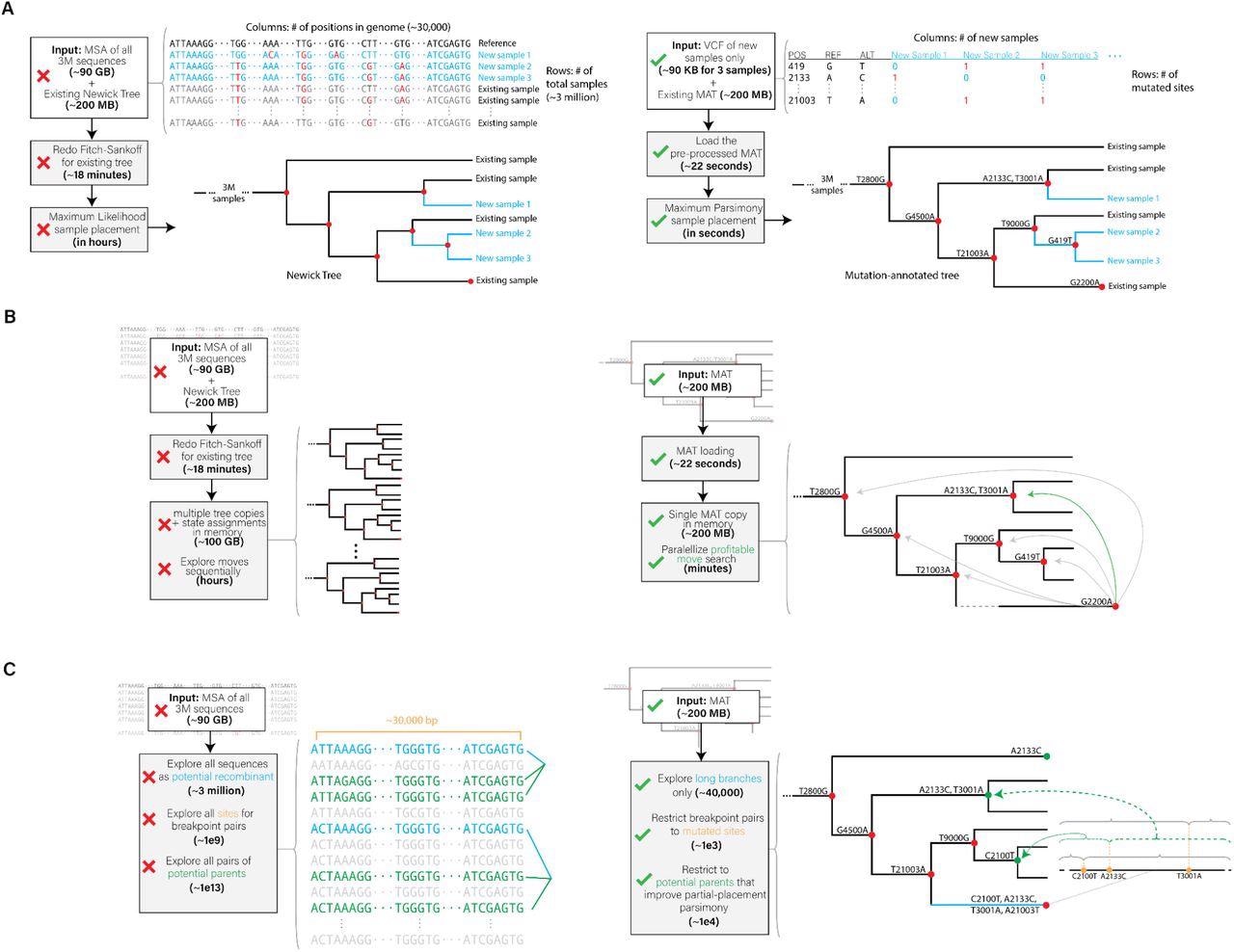 Innovative optimizations realized in (A) UShER, (B) matOptimize and (C) RIPPLES for phylogenetic placement, tree optimization and recombination detection, respectively. The left side shows a representative illustration of the prior approaches and the right side illustrates the approach used in our tools.
Innovative optimizations realized in (A) UShER, (B) matOptimize and (C) RIPPLES for phylogenetic placement, tree optimization and recombination detection, respectively. The left side shows a representative illustration of the prior approaches and the right side illustrates the approach used in our tools.
Performance results of UShER, matOptimize, and RIPPLES
Speedup analysis highlighted the magnitude of improvement in runtime and peak memory that this phylogenetic package achieves relative to state-of-the-art tools. For phylogenetic placement, as compared to IQ-TREE2, UShER achieved 1439-fold speedup and 1300-fold improved memory efficiency, as well as placed 1000 new samples on the 100K-sample tree in just 15.4 seconds using 92 MB of RAM.
For tree optimization, as compared to TNT, matOptimize completed its optimization in just over one hour and remained more parsimony-optimal even after 24 hours. For recombination detection, placing a new sample on the 1M-sample tree using UShER and flagging it as a recombinant using RIPPLES took 35.65 seconds on average, which enabled real-time monitoring of the virus for recombination.
UShER maintained a strong scaling efficiency of over 85% in placing 100K new samples on the 1M-sample tree until 512 vCPUs were used, after which it dropped to 72.6% at 1024 vCPUs.
For matOptimize, its strong scaling efficiency rapidly deteriorated with parallelism. For instance, with 1024 vCPUs, the entire matOptimize run required only 11.5 minutes, with the parallel search phase requiring 7.5 minutes in total and less than 1.5 minutes on each iteration.
The authors anticipate improvement in strong scaling efficiency as the tree grows. RIPPLES achieved a strong scaling efficiency of over 80%, the highest of all programs, for comprehensively detecting recombinants from the 1M-sample tree at all parallelism levels. All the tools showed weak scaling efficiency of above 70%, as determined during weak scaling analysis.
Conclusions
The current study addressed the unmet needs imposed by the COVID-19 pandemic and developed a phylogenetic package for comprehensive phylogenetic analyses of SARS-CoV-2. COVID-19 phylogenetics has been crucial for genomic surveillance of SARS-CoV-2 and its variants, as well as for their identification and naming, thus supporting their potential relevance in epidemiological studies.
This tool, therefore, helps in estimating the reproduction number (R0) of the SARS-CoV-2 or its particular variant. In addition, phylogenetics may establish transmission links between seemingly unrelated SARS-CoV-2 infections.
Of all the programs of the phylogenetic package, UShER and RIPPLES showed the potential to empower individual research labs to incorporate their SARS-CoV-2 genomic sequences onto a global phylogeny, discover evidence for recombination from a massive search space, and subsequently provide a real-time response. RIPPLES could also be used in high-performance computing (HPC) setting to detect recombination events from the vast SARS-CoV-2 phylogeny within a few hours. With matUtils, it was possible to rapidly query and visualize massive SARS-CoV-2 phylogenies.
Overall, these tools showed the potential to empower the global scientific community to study the SARS-CoV-2 evolution and transmission at an extraordinary scale, resolution, and speed.

 This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
This news article was a review of a preliminary scientific report that had not undergone peer-review at the time of publication. Since its initial publication, the scientific report has now been peer reviewed and accepted for publication in a Scientific Journal. Links to the preliminary and peer-reviewed reports are available in the Sources section at the bottom of this article. View Sources
Journal references:
- Preliminary scientific report.
Ye, C., Thornlow, B., Kramer, A., et al. (2021). Pandemic-scale phylogenetics. bioRxiv. doi:10.1101/2021.12.03.470766. https://www.biorxiv.org/content/10.1101/2021.12.03.470766v1.
- Peer reviewed and published scientific report.
Turakhia, Yatish, Bryan Thornlow, Angie Hinrichs, Jakob McBroome, Nicolas Ayala, Cheng Ye, Kyle Smith, et al. 2022. “Pandemic-Scale Phylogenomics Reveals the SARS-CoV-2 Recombination Landscape.” Nature 609 (7929): 994–97. https://doi.org/10.1038/s41586-022-05189-9. https://www.nature.com/articles/s41586-022-05189-9.